|
|
Analogs
-
4563258
-
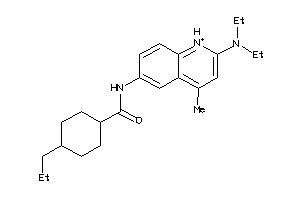
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-1-E |
Glucosylceramidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
514 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
11.97 |
-26.44 |
2 |
4 |
1 |
46 |
354.518 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.03 |
11.61 |
-11.14 |
1 |
4 |
0 |
45 |
353.51 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
3649803
-
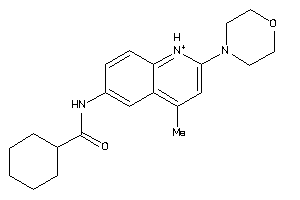
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-1-E |
Glucosylceramidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
55 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.37 |
11.07 |
-30.1 |
2 |
5 |
1 |
56 |
396.555 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.37 |
10.75 |
-13.74 |
1 |
5 |
0 |
54 |
395.547 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-1-E |
Glucosylceramidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
168 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
5.8 |
-15.38 |
2 |
7 |
0 |
101 |
371.418 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-1-E |
Glucosylceramidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8440 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.84 |
7.17 |
-23.36 |
1 |
7 |
0 |
92 |
402.501 |
6 |
↓
|
|
|
|
Analogs
-
5184768
-
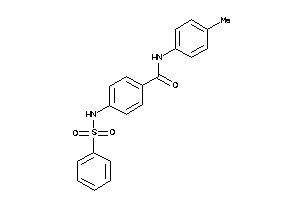
-
22688946
-
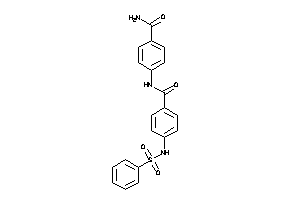
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-1-E |
Glucosylceramidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6460 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
6.45 |
-16.01 |
2 |
5 |
0 |
75 |
352.415 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.59 |
6.47 |
-50.33 |
1 |
5 |
-1 |
77 |
351.407 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-1-E |
Glucosylceramidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7150 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
5.09 |
-14.86 |
2 |
5 |
0 |
75 |
332.425 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.33 |
5.11 |
-49.38 |
1 |
5 |
-1 |
77 |
331.417 |
7 |
↓
|
|
|
|
Analogs
-
4563123
-
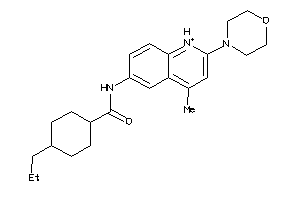
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-1-E |
Glucosylceramidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
31 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
9.19 |
-30.82 |
2 |
5 |
1 |
56 |
354.474 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.36 |
8.89 |
-13.58 |
1 |
5 |
0 |
54 |
353.466 |
3 |
↓
|
|
|
|
Analogs
-
25449721
-
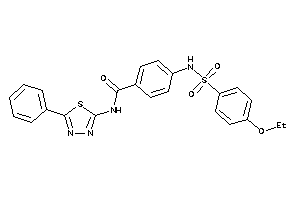
-
25449729
-
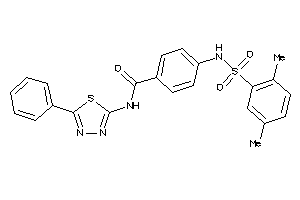
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-1-E |
Glucosylceramidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
5.64 |
-22.21 |
2 |
7 |
0 |
101 |
388.474 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.60 |
5.66 |
-54.92 |
1 |
7 |
-1 |
103 |
387.466 |
6 |
↓
|
|
|
|
Analogs
-
4563252
-
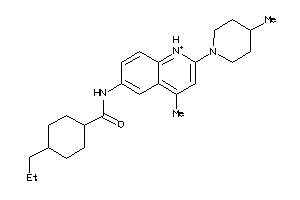
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-1-E |
Glucosylceramidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
268 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.43 |
12.69 |
-28.13 |
2 |
4 |
1 |
46 |
380.556 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.43 |
12.41 |
-12.79 |
1 |
4 |
0 |
45 |
379.548 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-1-E |
Glucosylceramidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
870 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
10 |
-10.98 |
3 |
8 |
0 |
95 |
364.837 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.61 |
10.42 |
-24.27 |
4 |
8 |
1 |
97 |
365.845 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.61 |
10.43 |
-25.48 |
4 |
8 |
1 |
97 |
365.845 |
7 |
↓
|
|
|
|
|
|
|
|