|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
736 |
0.45 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
713 |
0.45 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
96 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
9.46 |
-5.82 |
1 |
3 |
0 |
38 |
318.149 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
116 |
0.51 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
398 |
0.47 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
256 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.09 |
9.9 |
-5.54 |
1 |
3 |
0 |
38 |
334.604 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
330 |
0.50 |
Binding ≤ 10μM
|
|
GRM5-2-E |
Metabotropic Glutamate Receptor 5 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
5330 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.71 |
7.18 |
-18.33 |
1 |
4 |
0 |
66 |
237.262 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.71 |
6.98 |
-36.17 |
2 |
4 |
1 |
67 |
238.27 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1530 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
7.11 |
-13.29 |
1 |
3 |
0 |
42 |
267.115 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1080 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
7.88 |
-11.33 |
1 |
3 |
0 |
42 |
338.148 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.04 |
7.68 |
-27.1 |
2 |
3 |
1 |
43 |
339.156 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5490 |
0.46 |
Binding ≤ 10μM
|
|
HXK4-1-E |
Hexokinase Type IV (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.76 |
-0.78 |
-7.48 |
1 |
3 |
0 |
41 |
252.726 |
2 |
↓
|
|
|
|
Analogs
-
6234298
-
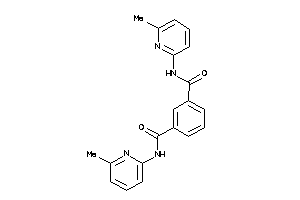
-
23082096
-
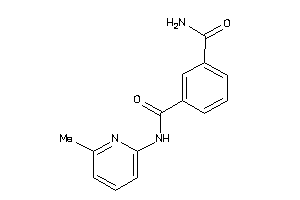
-
496676
-
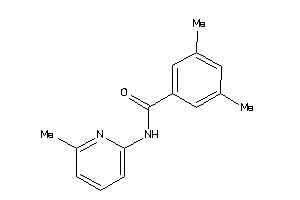
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1840 |
0.47 |
Binding ≤ 10μM
|
|
GRM5-2-E |
Metabotropic Glutamate Receptor 5 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
5900 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
7.45 |
-14.22 |
1 |
3 |
0 |
42 |
226.279 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.41 |
7.25 |
-27.83 |
2 |
3 |
1 |
43 |
227.287 |
2 |
↓
|
|
|
|
Analogs
-
13676104
-
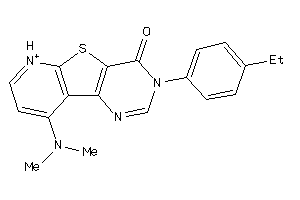
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.48 |
Binding ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
500 |
0.37 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.48 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
890 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
10.48 |
-39.44 |
1 |
5 |
1 |
52 |
337.428 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.97 |
10.05 |
-14.15 |
0 |
5 |
0 |
51 |
336.42 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1030 |
0.35 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
16 |
0.45 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1810 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.20 |
-1.46 |
-41.13 |
1 |
5 |
1 |
52 |
357.846 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
61 |
0.56 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Functional ≤ 10μM
|
|
GRM5-2-E |
Metabotropic Glutamate Receptor 5 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
38 |
0.58 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.41 |
2.91 |
-51.9 |
1 |
6 |
-1 |
80 |
265.68 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3070 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.14 |
5.26 |
-13.55 |
1 |
5 |
0 |
60 |
216.244 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
190 |
0.50 |
Binding ≤ 10μM
|
|
GRM5-2-E |
Metabotropic Glutamate Receptor 5 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
73 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
2.65 |
-13.24 |
0 |
6 |
0 |
80 |
248.249 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
650 |
0.51 |
Binding ≤ 10μM
|
|
GRM5-2-E |
Metabotropic Glutamate Receptor 5 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
5480 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
-0.43 |
-11.66 |
1 |
3 |
0 |
41 |
291.148 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.77 |
-0.78 |
-27.4 |
2 |
3 |
1 |
43 |
292.156 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1730 |
0.47 |
Binding ≤ 10μM
|
|
GRM5-2-E |
Metabotropic Glutamate Receptor 5 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1870 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
0.18 |
-11.79 |
1 |
3 |
0 |
42 |
246.697 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.64 |
-0.17 |
-27.49 |
2 |
3 |
1 |
43 |
247.705 |
2 |
↓
|
|
|
|
Analogs
-
184798
-
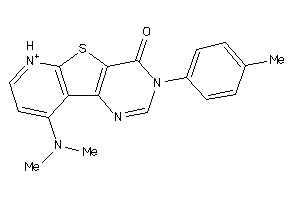
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.46 |
Binding ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
194 |
0.38 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
442 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
9.72 |
-40.51 |
1 |
5 |
1 |
52 |
351.455 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
350 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.51 |
10.45 |
-13.88 |
0 |
4 |
0 |
33 |
344.842 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
134 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.10 |
2.46 |
-8.98 |
3 |
7 |
0 |
97 |
292.73 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
254 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.26 |
3.05 |
-10.37 |
1 |
5 |
0 |
68 |
234.646 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.65 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.07 |
6.52 |
-5.15 |
0 |
3 |
0 |
46 |
279.126 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3900 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
3.92 |
-20.54 |
3 |
5 |
0 |
78 |
287.706 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3900 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
7.18 |
-12.56 |
2 |
3 |
0 |
45 |
270.719 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3536 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.07 |
1.74 |
-11.76 |
3 |
6 |
0 |
94 |
215.216 |
2 |
↓
|
|
|
|
Analogs
-
36882902
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7580 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.24 |
6.89 |
-13.83 |
1 |
3 |
0 |
42 |
248.232 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
0.87 |
-14.74 |
1 |
3 |
0 |
41 |
288.35 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.95 |
0.51 |
-30.04 |
2 |
3 |
1 |
46 |
289.358 |
2 |
↓
|
|