|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 113 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH1-2-E |
Histamine H1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
190 |
1.18 |
Binding ≤ 10μM
|
|
HRH2-2-E |
Histamine H2 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1200 |
1.04 |
Binding ≤ 10μM
|
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
77 |
1.24 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
75 |
1.25 |
Binding ≤ 10μM
|
|
HRH1-2-E |
Histamine H1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
45 |
1.29 |
Functional ≤ 10μM
|
|
HRH2-1-E |
Histamine H2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
1.08 |
Functional ≤ 10μM
|
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
88 |
1.23 |
Functional ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1800 |
1.01 |
Functional ≤ 10μM
|
|
Z50512-3-O |
Cavia Porcellus (cluster #3 Of 7), Other |
Other |
980 |
1.05 |
Functional ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 12), Other |
Other |
62 |
1.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.85 |
0.85 |
-42.34 |
4 |
3 |
1 |
56 |
112.156 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.85 |
1.31 |
-96.9 |
5 |
3 |
2 |
58 |
113.164 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
85 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
8.71 |
-42.22 |
5 |
7 |
1 |
104 |
260.325 |
6 |
↓
|
|
Ref
Reference (pH 7)
|
0.76 |
8.94 |
-39.38 |
5 |
7 |
1 |
104 |
260.325 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.76 |
8.45 |
-14.39 |
4 |
7 |
0 |
102 |
259.317 |
6 |
↓
|
|
|
|
|
|
|
Analogs
-
39125091
-
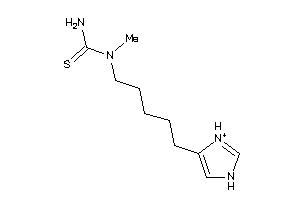
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH2-2-E |
Histamine H2 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3981 |
0.54 |
Binding ≤ 10μM
|
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.71 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
40 |
0.74 |
Binding ≤ 10μM
|
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.86 |
5.09 |
-44.36 |
4 |
4 |
1 |
54 |
213.33 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.86 |
4.6 |
-17.89 |
3 |
4 |
0 |
53 |
212.322 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.86 |
4.58 |
-18.25 |
3 |
4 |
0 |
53 |
212.322 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.54 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.54 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
9.95 |
-84.83 |
5 |
4 |
2 |
68 |
416.332 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.48 |
9.46 |
-41.88 |
4 |
4 |
1 |
66 |
415.324 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.35 |
9.37 |
-37.46 |
4 |
4 |
1 |
69 |
415.324 |
8 |
↓
|
|
|
|
Analogs
-
44411624
-
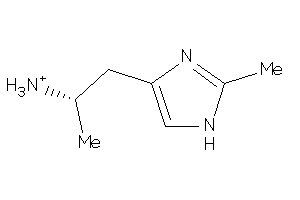
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
794 |
0.95 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.76 |
0.7 |
-42.42 |
4 |
3 |
1 |
56 |
126.183 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.76 |
1.12 |
-94.52 |
5 |
3 |
2 |
58 |
127.191 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
9.33 |
-42.73 |
2 |
4 |
1 |
56 |
271.34 |
7 |
↓
|
|
Ref
Reference (pH 7)
|
2.69 |
8.84 |
-13.94 |
1 |
4 |
0 |
55 |
270.332 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.69 |
9.35 |
-42.76 |
2 |
4 |
1 |
56 |
271.34 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
8.47 |
-84.45 |
5 |
4 |
2 |
70 |
310.854 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.67 |
8.11 |
-8.6 |
3 |
4 |
0 |
65 |
308.838 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.67 |
8.13 |
-38.57 |
4 |
4 |
1 |
66 |
309.846 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.97 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1995 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
6.99 |
-83.7 |
3 |
3 |
2 |
34 |
181.283 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.15 |
6.43 |
-36.78 |
2 |
3 |
1 |
33 |
180.275 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
1.05 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
22 |
0.89 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
0.55 |
4.31 |
-43.95 |
3 |
3 |
1 |
45 |
166.248 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.75 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.64 |
Binding ≤ 10μM
|
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.67 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
7.82 |
-35.3 |
2 |
3 |
1 |
39 |
217.292 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.19 |
7.33 |
-9.75 |
1 |
3 |
0 |
38 |
216.284 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.19 |
7.31 |
-9.2 |
1 |
3 |
0 |
38 |
216.284 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
1.11 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6 |
1.05 |
Binding ≤ 10μM
|
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
1.06 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.05 |
2.31 |
-83.11 |
6 |
4 |
2 |
82 |
172.257 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.05 |
1.84 |
-31.45 |
5 |
4 |
1 |
80 |
171.249 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1259 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.14 |
4.72 |
-46 |
4 |
3 |
1 |
56 |
256.251 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.14 |
5.13 |
-106.57 |
5 |
3 |
2 |
58 |
257.259 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
126 |
0.48 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
347 |
0.45 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Binding ≤ 10μM
|
|
HRH2-2-E |
Histamine H2 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Binding ≤ 10μM
|
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.50 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
63 |
0.50 |
Binding ≤ 10μM
|
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Functional ≤ 10μM |
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
680 |
0.43 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6095 |
0.37 |
ADME/T ≤ 10μM
|
|
Z50425-12-O |
Plasmodium Falciparum (cluster #12 Of 22), Other |
Other |
10000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
9.81 |
-47.23 |
3 |
4 |
1 |
45 |
293.46 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.66 |
9.82 |
-46.74 |
3 |
4 |
1 |
45 |
293.46 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.66 |
9.19 |
-13.36 |
2 |
4 |
0 |
44 |
292.452 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
39258812
-
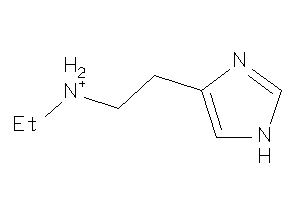
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
1.31 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
316 |
1.01 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.13 |
2.72 |
-37.82 |
3 |
3 |
1 |
45 |
126.183 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.13 |
3.19 |
-92.89 |
4 |
3 |
2 |
47 |
127.191 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH2-2-E |
Histamine H2 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7943 |
0.79 |
Binding ≤ 10μM
|
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6310 |
0.81 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
1.14 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.63 |
1.35 |
-44 |
4 |
3 |
1 |
56 |
126.183 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.63 |
1.82 |
-96.48 |
5 |
3 |
2 |
58 |
127.191 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH2-1-E |
Histamine H2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6310 |
0.73 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5012 |
0.74 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.08 |
0.12 |
-49.05 |
5 |
3 |
1 |
67 |
158.25 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.08 |
0.57 |
-91.2 |
6 |
3 |
2 |
68 |
159.258 |
2 |
↓
|
|
|
|
Analogs
-
39133125
-
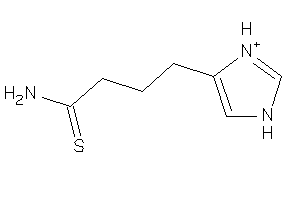
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
1.06 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
251 |
0.84 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.20 |
3.65 |
-84.48 |
5 |
3 |
2 |
58 |
155.245 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.20 |
3.16 |
-46.83 |
4 |
3 |
1 |
56 |
154.237 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
1.05 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
251 |
0.77 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.84 |
5.18 |
-39.12 |
2 |
3 |
1 |
43 |
160.2 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
0.84 |
5.21 |
-39.09 |
2 |
3 |
1 |
43 |
160.2 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.84 |
4.7 |
-9.63 |
1 |
3 |
0 |
42 |
159.192 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
1.14 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
32 |
1.17 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.58 |
2.09 |
-91.44 |
5 |
3 |
2 |
58 |
127.191 |
3 |
↓
|
|