|
|
Analogs
-
4201359
-
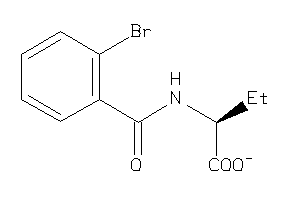
-
4201360
-
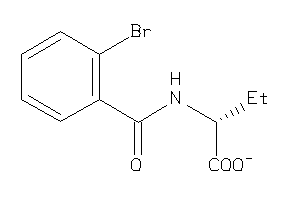
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3840 |
0.45 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3840 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.60 |
2.1 |
-60.58 |
2 |
5 |
-1 |
89 |
301.116 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
1.61 |
-25.31 |
0 |
7 |
0 |
86 |
453.564 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.53 |
Binding ≤ 10μM
|
|
LYAM2-1-E |
Selectin E (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
5.65 |
-9.48 |
2 |
3 |
0 |
56 |
300.408 |
3 |
↓
|
|
|
|
Analogs
-
3369682
-
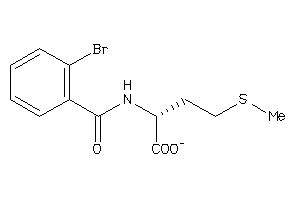
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7460 |
0.40 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7460 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.46 |
-1.16 |
-56.54 |
1 |
4 |
-1 |
69 |
331.211 |
6 |
↓
|
|
|
|
|
|
|
Analogs
-
5280396
-
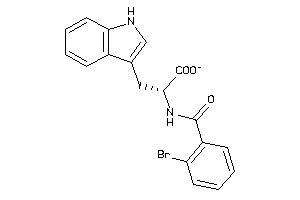
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1720 |
0.34 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1720 |
0.34 |
Binding ≤ 10μM
|
|
ITB2-1-E |
Integrin Beta-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1720 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.62 |
-1.45 |
-60.75 |
2 |
5 |
-1 |
85 |
386.225 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1080 |
0.46 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1080 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.11 |
1.41 |
-60.86 |
3 |
6 |
-1 |
112 |
314.115 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
9863973
-
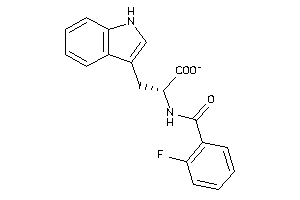
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7400 |
0.30 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7400 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.98 |
-0.11 |
-65.13 |
2 |
5 |
-1 |
85 |
325.319 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
31685399
-
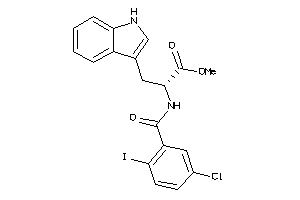
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8900 |
0.29 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8900 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
8.66 |
-61.09 |
2 |
5 |
-1 |
85 |
433.225 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.37 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
250 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.61 |
7.97 |
-66.35 |
2 |
5 |
-1 |
85 |
359.764 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8400 |
0.28 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8400 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
8.61 |
-58.9 |
2 |
5 |
-1 |
85 |
376.219 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.51 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.12 |
1.05 |
-67.09 |
3 |
6 |
-1 |
112 |
287.654 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.45 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.83 |
4.82 |
-60.26 |
2 |
6 |
-1 |
98 |
294.237 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.83 |
5.33 |
-70.87 |
3 |
6 |
0 |
99 |
295.245 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.83 |
5.91 |
-71.25 |
2 |
6 |
-1 |
98 |
294.237 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.49 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
230 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.64 |
0.67 |
-70.03 |
3 |
6 |
-1 |
112 |
271.199 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.31 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.26 |
8.24 |
-62.72 |
2 |
5 |
-1 |
85 |
321.356 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.34 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.49 |
9.07 |
-60.55 |
2 |
5 |
-1 |
85 |
341.774 |
5 |
↓
|
|
|
|
Analogs
-
21797839
-
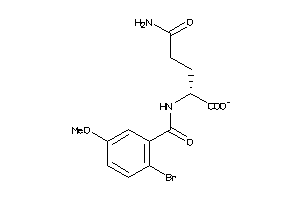
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1770 |
0.42 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1770 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.06 |
2.05 |
-63.5 |
3 |
6 |
-1 |
112 |
328.142 |
6 |
↓
|
|
|
|
|
|
|
Analogs
-
12031033
-
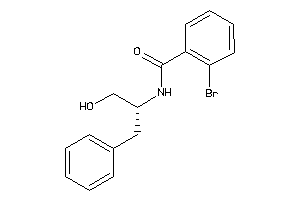
-
12031037
-
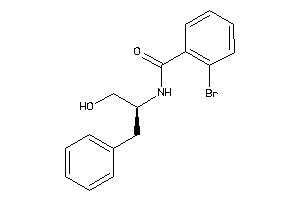
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4890 |
0.35 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4890 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
-0.5 |
-58.74 |
1 |
4 |
-1 |
69 |
347.188 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4260 |
0.47 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4260 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.01 |
4.62 |
-52.4 |
1 |
4 |
-1 |
69 |
303.157 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.01 |
5.16 |
-116.46 |
1 |
4 |
-2 |
69 |
302.149 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1090 |
0.38 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1090 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.99 |
5.38 |
-59.16 |
2 |
5 |
-1 |
89 |
363.187 |
5 |
↓
|
|
|
|
Analogs
-
4201359
-

-
4201360
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICAM1-1-E |
Intercellular Adhesion Molecule-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6150 |
0.46 |
Binding ≤ 10μM
|
|
ITAL-1-E |
Integrin Alpha-L (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6150 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.96 |
1.05 |
-60.06 |
2 |
5 |
-1 |
89 |
287.089 |
4 |
↓
|
|