|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA4-1-E |
Integrin Alpha-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.36 |
Binding ≤ 10μM
|
|
ITB7-1-E |
Integrin Beta-7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
-0.01 |
-9.49 |
1 |
5 |
0 |
71 |
292.294 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA4-1-E |
Integrin Alpha-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.32 |
Binding ≤ 10μM
|
|
ITB7-1-E |
Integrin Beta-7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
2.17 |
-10.39 |
1 |
5 |
0 |
71 |
360.291 |
3 |
↓
|
|
|
|
Analogs
-
2181582
-
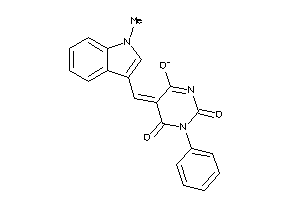
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA4-1-E |
Integrin Alpha-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4100 |
0.29 |
Binding ≤ 10μM
|
|
ITB7-1-E |
Integrin Beta-7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4100 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
6.05 |
-54.07 |
0 |
6 |
-1 |
80 |
344.35 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.85 |
8.7 |
-11.41 |
1 |
6 |
0 |
77 |
345.358 |
2 |
↓
|
|
|
|
Analogs
-
6926619
-
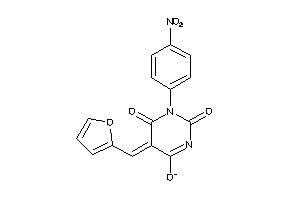
-
9109556
-
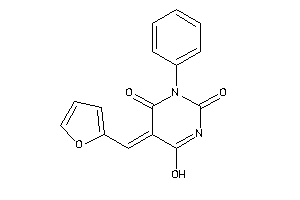
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA4-1-E |
Integrin Alpha-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.39 |
Binding ≤ 10μM
|
|
ITB7-1-E |
Integrin Beta-7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
4.57 |
-49.77 |
0 |
6 |
-1 |
88 |
281.247 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.89 |
6.36 |
-12.63 |
1 |
6 |
0 |
85 |
282.255 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA4-1-E |
Integrin Alpha-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
180 |
0.31 |
Binding ≤ 10μM
|
|
ITB7-1-E |
Integrin Beta-7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
180 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.68 |
9.65 |
-57.28 |
0 |
6 |
-1 |
80 |
394.41 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.22 |
11.43 |
-14.63 |
1 |
6 |
0 |
77 |
395.418 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA4-1-E |
Integrin Alpha-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.31 |
Binding ≤ 10μM
|
|
ITB7-1-E |
Integrin Beta-7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
3.79 |
-61.75 |
2 |
10 |
-1 |
143 |
432.453 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA4-1-E |
Integrin Alpha-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
60 |
0.35 |
Binding ≤ 10μM
|
|
ITB7-1-E |
Integrin Beta-7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
60 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
7.11 |
-57.96 |
1 |
6 |
-1 |
91 |
380.383 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.16 |
9.71 |
-14.67 |
2 |
6 |
0 |
88 |
381.391 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA4-1-E |
Integrin Alpha-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
|
ITB7-1-E |
Integrin Beta-7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
7.01 |
-10.59 |
2 |
6 |
0 |
88 |
331.331 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
0.27 |
-63.3 |
1 |
6 |
-1 |
87 |
473.332 |
8 |
↓
|
|