|
|
Analogs
-
3875336
-
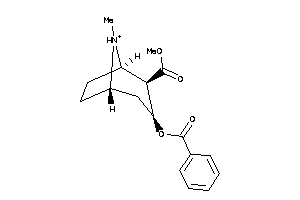
-
4214983
-
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
3.68 |
-33.02 |
1 |
5 |
1 |
57 |
318.393 |
6 |
↓
|
|
|
|
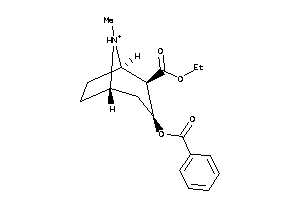
Analogs
-
4214983
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
3.43 |
-42.14 |
1 |
5 |
1 |
57 |
318.393 |
6 |
↓
|
|
|
|
Analogs
-
4214983
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-4-E |
HERG (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
7244 |
0.33 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9820 |
0.32 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
208 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
9.75 |
-41.49 |
1 |
5 |
1 |
57 |
304.366 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.87 |
7.41 |
-6.91 |
0 |
5 |
0 |
56 |
303.358 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-4-E |
HERG (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
8000 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
4.04 |
-65.82 |
1 |
6 |
0 |
83 |
510.512 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-4-E |
HERG (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
8913 |
0.54 |
Binding ≤ 10μM
|
|
Z104290-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #2 Of 4), Other |
Other |
8900 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.17 |
3.45 |
-5.36 |
0 |
3 |
0 |
30 |
181.235 |
2 |
↓
|
|
|
|
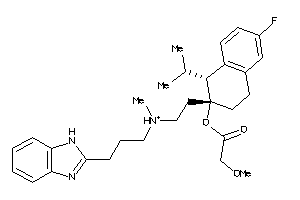
Analogs
-
607970
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1340 |
0.23 |
Binding ≤ 10μM
|
|
CAC1G-1-E |
Voltage-gated T-type Calcium Channel Alpha-1G Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
860 |
0.24 |
Binding ≤ 10μM |
|
CAC1H-1-E |
Voltage-gated T-type Calcium Channel Alpha-1H Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.23 |
Binding ≤ 10μM
|
|
CAC1I-1-E |
Voltage-gated T-type Calcium Channel Alpha-1I Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
126 |
0.27 |
Binding ≤ 10μM
|
|
KCNH2-4-E |
HERG (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
806 |
0.24 |
Binding ≤ 10μM
|
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
416 |
0.25 |
Functional ≤ 10μM
|
|
CAC1G-1-E |
Voltage-gated T-type Calcium Channel Alpha-1G Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
840 |
0.24 |
Functional ≤ 10μM
|
|
CAC1H-1-E |
Voltage-gated T-type Calcium Channel Alpha-1H Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.29 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1430 |
0.23 |
Functional ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.22 |
Functional ≤ 10μM
|
|
MDR3-2-E |
P-glycoprotein 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7400 |
0.20 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
130 |
0.27 |
ADME/T ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
1 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
398 |
0.25 |
Functional ≤ 10μM
|
|
Z80107-2-O |
COS-7 (Kidney Cells) (cluster #2 Of 2), Other |
Other |
1430 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.56 |
14.86 |
-54.6 |
2 |
6 |
1 |
69 |
496.647 |
12 |
↓
|
|
|
|
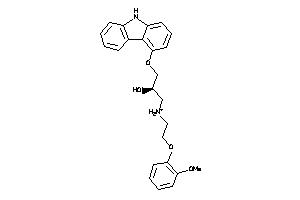
Analogs
-
1530580
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
7.38 |
-51.26 |
4 |
6 |
1 |
80 |
407.49 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.56 |
6.05 |
-14.85 |
3 |
6 |
0 |
76 |
406.482 |
10 |
↓
|
|
|
|
Analogs
-
1530579
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
7.36 |
-51.12 |
4 |
6 |
1 |
80 |
407.49 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.56 |
5.94 |
-15.03 |
3 |
6 |
0 |
76 |
406.482 |
10 |
↓
|
|