|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(2R,4aS,5R,10bS)-5-phenyl-9-(trifluoromethyl)-3,4,4a,5,6,10b-hexahydro-2H-pyrano[3,2-c]quinolin-2-y
[(2R,4aS,5R,10bS)-5-phenyl-9-(tr…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.41 |
Binding ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
4 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
7.3 |
-45.4 |
4 |
3 |
1 |
49 |
363.403 |
3 |
↓
|
|
|
|
Analogs
-
30780140
-
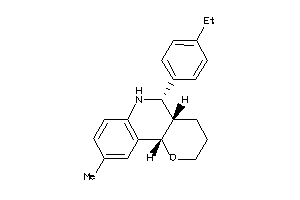
-
30780980
-
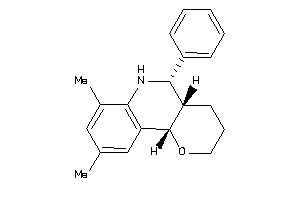
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
420 |
0.43 |
Binding ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
840 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
8.68 |
-4.14 |
1 |
2 |
0 |
21 |
279.383 |
1 |
↓
|
|
|
|
Analogs
-
30780184
-
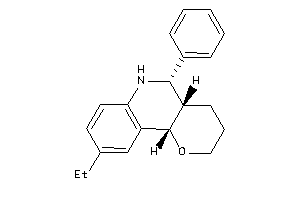
-
30780463
-
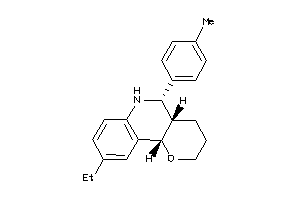
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aS,5R,10bS)-9-ethyl-5-phenyl-3,4,4a,5,6,10b-hexahydro-2H-pyrano[3,2-c]quinoline
(4aS,5R,10bS)-9-ethyl-5-phenyl-3…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
130 |
0.44 |
Binding ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
370 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.80 |
9.47 |
-3.99 |
1 |
2 |
0 |
21 |
293.41 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.41 |
Binding ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
1400 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.56 |
-1.35 |
-3.9 |
1 |
2 |
0 |
21 |
299.801 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.27 |
Binding ≤ 10μM |
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM |
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
3330 |
0.20 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
7020 |
0.19 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
940 |
0.22 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
2100 |
0.21 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
3000 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.94 |
2.26 |
-55.93 |
1 |
6 |
1 |
59 |
596.977 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.29 |
Binding ≤ 10μM
|
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.27 |
Binding ≤ 10μM |
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM |
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
3330 |
0.20 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
7020 |
0.19 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
940 |
0.22 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
2100 |
0.21 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
3000 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.94 |
17.46 |
-55.26 |
1 |
6 |
1 |
60 |
596.977 |
10 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
5.55 |
-15.6 |
3 |
4 |
0 |
61 |
302.399 |
1 |
↓
|
|
|
|
Analogs
-
36184686
-
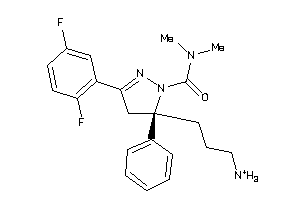
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
5 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.74 |
9.84 |
-50.56 |
3 |
5 |
1 |
64 |
387.454 |
5 |
↓
|
|
|
|
Analogs
-
34604066
-
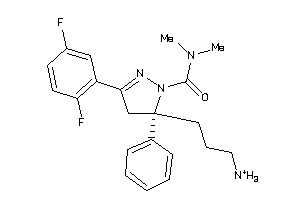
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.74 |
9.75 |
-52.61 |
3 |
5 |
1 |
64 |
387.454 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
480 |
0.37 |
Binding ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
2000 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
8.96 |
-10.98 |
1 |
4 |
0 |
49 |
362.52 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9900 |
0.30 |
Binding ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
7100 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
-0.56 |
-11.79 |
1 |
4 |
0 |
49 |
348.493 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4300 |
0.30 |
Binding ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
7200 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
-3.33 |
-11.09 |
1 |
5 |
0 |
58 |
376.503 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
-1.58 |
-11.68 |
1 |
4 |
0 |
49 |
334.466 |
3 |
↓
|
|
|
|
Analogs
-
643915
-
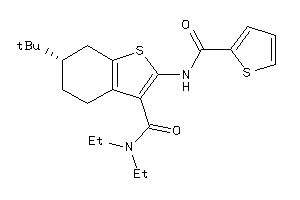
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5900 |
0.29 |
Binding ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
6100 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
-0.22 |
-10.96 |
1 |
4 |
0 |
49 |
376.547 |
5 |
↓
|
|
|
|
Analogs
-
643915
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5900 |
0.29 |
Binding ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
6100 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
-0.22 |
-11.08 |
1 |
4 |
0 |
49 |
376.547 |
5 |
↓
|
|
|
|
Analogs
-
4425506
-
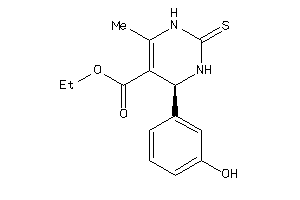
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7452 |
0.36 |
Binding ≤ 10μM |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6100 |
0.37 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
1500 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
5.18 |
-10.47 |
3 |
5 |
0 |
71 |
292.36 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.45 |
3.94 |
-38.16 |
2 |
5 |
-1 |
71 |
291.352 |
4 |
↓
|
|
|
|
Analogs
-
4425504
-
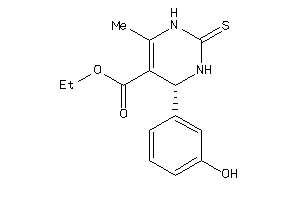
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7452 |
0.36 |
Binding ≤ 10μM |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Binding ≤ 10μM
|
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6100 |
0.37 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
1500 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
5.13 |
-10.78 |
3 |
5 |
0 |
71 |
292.36 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.45 |
3.93 |
-38.16 |
2 |
5 |
-1 |
71 |
291.352 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1649 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
1.85 |
-4.42 |
2 |
1 |
0 |
26 |
237.224 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.29 |
Binding ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
8600 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
-2.08 |
-10.63 |
2 |
6 |
0 |
76 |
389.455 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5800 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
-0.54 |
-11.16 |
1 |
4 |
0 |
49 |
376.547 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
540 |
0.35 |
Binding ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
5100 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.49 |
-2.22 |
-11.03 |
1 |
4 |
0 |
49 |
374.531 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.20 |
11.16 |
-15.75 |
3 |
6 |
0 |
82 |
458.415 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.20 |
11.33 |
-38.16 |
4 |
6 |
1 |
83 |
459.423 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.25 |
Binding ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8000 |
0.22 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
11.08 |
-15.64 |
3 |
6 |
0 |
82 |
440.425 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.10 |
11.25 |
-33.71 |
4 |
6 |
1 |
83 |
441.433 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.48 |
12.81 |
-15.09 |
3 |
5 |
0 |
73 |
456.493 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.48 |
12.98 |
-34.05 |
4 |
5 |
1 |
74 |
457.501 |
7 |
↓
|
|