|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA-2-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM |
|
KAPCA-2-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Binding ≤ 10μM
|
|
KAPCB-2-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM |
|
KAPCB-2-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM |
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Binding ≤ 10μM
|
|
KGP1-1-E |
CGMP-dependent Protein Kinase 1 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5800 |
0.37 |
Binding ≤ 10μM
|
|
KGP2-1-E |
CGMP-dependent Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5800 |
0.37 |
Binding ≤ 10μM
|
|
KPCA-6-E |
Protein Kinase C Alpha (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCA-6-E |
Protein Kinase C Alpha (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCB-3-E |
Protein Kinase C Beta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCB-3-E |
Protein Kinase C Beta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD-4-E |
Protein Kinase C Delta (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD-4-E |
Protein Kinase C Delta (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCE-4-E |
Protein Kinase C Epsilon (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCE-4-E |
Protein Kinase C Epsilon (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCG-2-E |
Protein Kinase C Gamma (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCG-2-E |
Protein Kinase C Gamma (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCL-3-E |
Protein Kinase C Eta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCL-3-E |
Protein Kinase C Eta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCZ-4-E |
Protein Kinase C Zeta (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCZ-4-E |
Protein Kinase C Zeta (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
Q95J97-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5400 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-1-B |
Beta-lactamase (cluster #1 Of 6), Bacterial |
Bacteria |
8000 |
0.30 |
Binding ≤ 10μM
|
|
AMPH-1-B |
Penicillin-binding Protein AmpH (cluster #1 Of 2), Bacterial |
Bacteria |
8000 |
0.30 |
Binding ≤ 10μM
|
|
MDH-1-B |
Malate Dehydrogenase (cluster #1 Of 1), Bacterial |
Bacteria |
8000 |
0.30 |
Binding ≤ 10μM
|
|
CTRB1-3-E |
Beta-chymotrypsin (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.29 |
Binding ≤ 10μM
|
|
GLI1-1-E |
Zinc Finger Protein GLI1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.34 |
Binding ≤ 10μM
|
|
GLI2-1-E |
Zinc Finger Protein GLI2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.32 |
Binding ≤ 10μM
|
|
JAK3-2-E |
Tyrosine-protein Kinase JAK3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
61 |
0.42 |
Binding ≤ 10μM
|
|
KCC2A-1-E |
CaM Kinase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
184 |
0.39 |
Binding ≤ 10μM
|
|
KCC2B-1-E |
CaM Kinase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
184 |
0.39 |
Binding ≤ 10μM
|
|
KCC2D-1-E |
CaM Kinase II Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
184 |
0.39 |
Binding ≤ 10μM
|
|
KCC2G-1-E |
CaM Kinase II Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
184 |
0.39 |
Binding ≤ 10μM
|
|
KGP1-1-E |
CGMP-dependent Protein Kinase 1 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
650 |
0.36 |
Binding ≤ 10μM |
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
680 |
0.36 |
Binding ≤ 10μM |
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
680 |
0.36 |
Binding ≤ 10μM |
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
680 |
0.36 |
Binding ≤ 10μM |
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
680 |
0.36 |
Binding ≤ 10μM |
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2450 |
0.33 |
Binding ≤ 10μM |
|
M3K11-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
49 |
0.43 |
Binding ≤ 10μM
|
|
M3K9-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 9 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
138 |
0.40 |
Binding ≤ 10μM
|
|
MYLK-1-E |
Myosin Light Chain Kinase, Smooth Muscle (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.32 |
Binding ≤ 10μM |
|
Z104295-1-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #1 Of 2), Other |
Other |
830 |
0.35 |
Binding ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 7), Other |
Other |
4800 |
0.31 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
4000 |
0.31 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
1720 |
0.34 |
Functional ≤ 10μM
|
|
Z81024-8-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #8 Of 8), Other |
Other |
3200 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
6.35 |
-15.15 |
3 |
4 |
0 |
61 |
311.344 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3100 |
0.31 |
Binding ≤ 10μM
|
|
KCC2D-1-E |
CaM Kinase II Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1190 |
0.33 |
Binding ≤ 10μM
|
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
190 |
0.38 |
Binding ≤ 10μM
|
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM
|
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
190 |
0.38 |
Binding ≤ 10μM
|
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
190 |
0.38 |
Binding ≤ 10μM
|
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
190 |
0.38 |
Binding ≤ 10μM
|
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7516 |
0.29 |
Binding ≤ 10μM
|
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM
|
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
190 |
0.38 |
Binding ≤ 10μM
|
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM
|
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
190 |
0.38 |
Binding ≤ 10μM
|
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.42 |
Binding ≤ 10μM
|
|
SIRT2-3-E |
NAD-dependent Deacetylase Sirtuin 2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4700 |
0.30 |
Binding ≤ 10μM
|
|
Z104295-1-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #1 Of 2), Other |
Other |
5810 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
-2.53 |
-11.9 |
3 |
5 |
0 |
81 |
327.343 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-2-E |
G2/mitotic-specific Cyclin B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1130 |
0.33 |
Binding ≤ 10μM
|
|
CCNB2-2-E |
G2/mitotic-specific Cyclin B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1130 |
0.33 |
Binding ≤ 10μM
|
|
CCNB3-2-E |
G2/mitotic-specific Cyclin B3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1130 |
0.33 |
Binding ≤ 10μM
|
|
CCNE1-2-E |
G1/S-specific Cyclin E1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
|
CCNE2-2-E |
G1/S-specific Cyclin E2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1130 |
0.33 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
|
KCC2A-1-E |
CaM Kinase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.43 |
Binding ≤ 10μM
|
|
KCC2B-1-E |
CaM Kinase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.43 |
Binding ≤ 10μM
|
|
KCC2D-1-E |
CaM Kinase II Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.43 |
Binding ≤ 10μM
|
|
KCC2G-1-E |
CaM Kinase II Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.43 |
Binding ≤ 10μM
|
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
550 |
0.35 |
Binding ≤ 10μM |
|
Z104295-1-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #1 Of 2), Other |
Other |
163 |
0.38 |
Binding ≤ 10μM
|
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
200 |
0.38 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
4000 |
0.30 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
850 |
0.34 |
Functional ≤ 10μM
|
|
Z81024-8-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #8 Of 8), Other |
Other |
590 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
5.67 |
-11.48 |
3 |
5 |
0 |
82 |
325.327 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.32 |
Binding ≤ 10μM |
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.32 |
Binding ≤ 10μM |
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.32 |
Binding ≤ 10μM |
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM |
|
KPCB-3-E |
Protein Kinase C Beta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM |
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM |
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM |
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM |
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM |
|
KPCG-2-E |
Protein Kinase C Gamma (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM |
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM |
|
KPCL-3-E |
Protein Kinase C Eta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM |
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM |
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
9.35 |
-10.83 |
1 |
5 |
0 |
60 |
327.343 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.01 |
7.59 |
-45.67 |
0 |
5 |
-1 |
63 |
326.335 |
2 |
↓
|
|
|
|
Analogs
-
34372551
-
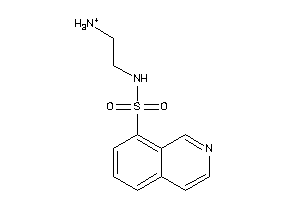
-
41661539
-
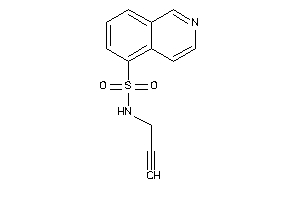
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA-2-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.47 |
Binding ≤ 10μM
|
|
KAPCB-2-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.47 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.47 |
Binding ≤ 10μM
|
|
KPCA-6-E |
Protein Kinase C Alpha (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
KPCB-3-E |
Protein Kinase C Beta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
KPCD-4-E |
Protein Kinase C Delta (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
KPCE-4-E |
Protein Kinase C Epsilon (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
KPCG-2-E |
Protein Kinase C Gamma (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
KPCL-3-E |
Protein Kinase C Eta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
KPCZ-4-E |
Protein Kinase C Zeta (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
P90584-2-E |
Protein Kinase Pfmrk (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.12 |
-0.55 |
-55.51 |
4 |
5 |
1 |
87 |
252.319 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.12 |
-0.94 |
-12.15 |
3 |
5 |
0 |
85 |
251.311 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA-2-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Binding ≤ 10μM
|
|
KAPCB-2-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Binding ≤ 10μM
|
|
KPCA-6-E |
Protein Kinase C Alpha (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCB-3-E |
Protein Kinase C Beta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD-4-E |
Protein Kinase C Delta (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCE-4-E |
Protein Kinase C Epsilon (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCG-2-E |
Protein Kinase C Gamma (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCL-3-E |
Protein Kinase C Eta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCZ-4-E |
Protein Kinase C Zeta (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-5-B |
Beta-lactamase (cluster #5 Of 6), Bacterial |
Bacteria |
5000 |
0.22 |
Binding ≤ 10μM
|
|
AMPH-1-B |
Penicillin-binding Protein AmpH (cluster #1 Of 2), Bacterial |
Bacteria |
5000 |
0.22 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
38 |
0.31 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.26 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.26 |
Binding ≤ 10μM
|
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
33 |
0.32 |
Binding ≤ 10μM
|
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM
|
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM |
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM |
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM |
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM |
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM
|
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM |
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM
|
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM |
|
KS6A3-1-E |
Ribosomal Protein S6 Kinase Alpha 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.34 |
Binding ≤ 10μM
|
|
KS6A5-1-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Binding ≤ 10μM
|
|
KS6B1-1-E |
Ribosomal Protein S6 Kinase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.33 |
Binding ≤ 10μM
|
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Binding ≤ 10μM
|
|
SIRT1-1-E |
NAD-dependent Deacetylase Sirtuin 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5100 |
0.22 |
Binding ≤ 10μM
|
|
SIRT2-3-E |
NAD-dependent Deacetylase Sirtuin 2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.26 |
Binding ≤ 10μM |
|
SIRT3-2-E |
NAD-dependent Deacetylase Sirtuin 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.23 |
Binding ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1960 |
0.24 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
840 |
0.26 |
Functional ≤ 10μM
|
|
Z81252-1-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #1 Of 11), Other |
Other |
1770 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
10.08 |
-46.88 |
5 |
7 |
1 |
111 |
458.567 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.47 |
10.2 |
-15.14 |
4 |
7 |
0 |
110 |
457.559 |
7 |
↓
|
|