|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LPAR1-1-E |
Lysophosphatidic Acid Receptor Edg-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
425 |
0.28 |
Binding ≤ 10μM
|
|
LPAR2-1-E |
Lysophosphatidic Acid Receptor Edg-4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1740 |
0.25 |
Binding ≤ 10μM
|
|
LPAR3-1-E |
Lysophosphatidic Acid Receptor Edg-7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1740 |
0.25 |
Binding ≤ 10μM
|
|
LPAR1-1-E |
Lysophosphatidic Acid Receptor Edg-2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
762 |
0.27 |
Functional ≤ 10μM
|
|
LPAR2-3-E |
Lysophosphatidic Acid Receptor Edg-4 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3384 |
0.24 |
Functional ≤ 10μM
|
|
LPAR3-3-E |
Lysophosphatidic Acid Receptor Edg-7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3384 |
0.24 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
3384 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.80 |
13.28 |
-48.06 |
1 |
7 |
-1 |
104 |
473.958 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.82 |
-3.88 |
-40.25 |
5 |
8 |
1 |
122 |
272.281 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LPAR1-1-E |
Lysophosphatidic Acid Receptor Edg-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
425 |
0.28 |
Binding ≤ 10μM
|
|
LPAR2-1-E |
Lysophosphatidic Acid Receptor Edg-4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1740 |
0.25 |
Binding ≤ 10μM
|
|
LPAR3-1-E |
Lysophosphatidic Acid Receptor Edg-7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1740 |
0.25 |
Binding ≤ 10μM
|
|
LPAR1-1-E |
Lysophosphatidic Acid Receptor Edg-2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
762 |
0.27 |
Functional ≤ 10μM
|
|
LPAR2-3-E |
Lysophosphatidic Acid Receptor Edg-4 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3384 |
0.24 |
Functional ≤ 10μM
|
|
LPAR3-3-E |
Lysophosphatidic Acid Receptor Edg-7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3384 |
0.24 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
3384 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.80 |
13.28 |
-48.01 |
1 |
7 |
-1 |
104 |
473.958 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.82 |
-2.1 |
-40.24 |
5 |
8 |
1 |
122 |
272.281 |
3 |
↓
|
|
|
|
Analogs
-
39899683
-
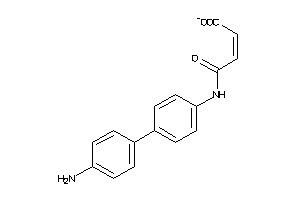
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LPAR2-4-E |
Lysophosphatidic Acid Receptor Edg-4 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
28 |
0.38 |
Functional ≤ 10μM
|
|
LPAR3-3-E |
Lysophosphatidic Acid Receptor Edg-7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7020 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.62 |
8.38 |
-94.92 |
2 |
8 |
-2 |
138 |
378.34 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LPAR2-4-E |
Lysophosphatidic Acid Receptor Edg-4 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.31 |
Functional ≤ 10μM
|
|
LPAR3-3-E |
Lysophosphatidic Acid Receptor Edg-7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
310 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.31 |
-0.63 |
-99 |
2 |
10 |
-2 |
156 |
486.436 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LPAR2-4-E |
Lysophosphatidic Acid Receptor Edg-4 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
355 |
0.36 |
Functional ≤ 10μM
|
|
LPAR3-3-E |
Lysophosphatidic Acid Receptor Edg-7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.70 |
2.89 |
-161.25 |
2 |
10 |
-3 |
179 |
345.243 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LPAR1-1-E |
Lysophosphatidic Acid Receptor Edg-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
48 |
0.34 |
Binding ≤ 10μM
|
|
LPAR3-1-E |
Lysophosphatidic Acid Receptor Edg-7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
230 |
0.31 |
Binding ≤ 10μM
|
|
LPAR5-1-E |
Lysophosphatidic Acid Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
292 |
0.30 |
Binding ≤ 10μM
|
|
LPAR1-1-E |
Lysophosphatidic Acid Receptor Edg-2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
94 |
0.33 |
Functional ≤ 10μM
|
|
LPAR3-3-E |
Lysophosphatidic Acid Receptor Edg-7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
752 |
0.29 |
Functional ≤ 10μM
|
|
LPAR5-2-E |
Lysophosphatidic Acid Receptor 5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
463 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
2.17 |
-46.27 |
0 |
9 |
-1 |
134 |
403.326 |
5 |
↓
|
|
|
|
Analogs
-
3212935
-
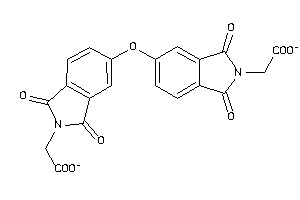
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LPAR1-1-E |
Lysophosphatidic Acid Receptor Edg-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
311 |
0.40 |
Binding ≤ 10μM
|
|
LPAR3-1-E |
Lysophosphatidic Acid Receptor Edg-7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1084 |
0.36 |
Binding ≤ 10μM
|
|
LPAR1-1-E |
Lysophosphatidic Acid Receptor Edg-2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
609 |
0.38 |
Functional ≤ 10μM
|
|
LPAR3-3-E |
Lysophosphatidic Acid Receptor Edg-7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2992 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.95 |
1.7 |
-52.89 |
0 |
6 |
-1 |
88 |
310.285 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LPAR3-3-E |
Lysophosphatidic Acid Receptor Edg-7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4504 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
1.92 |
-56.09 |
0 |
9 |
-1 |
134 |
341.255 |
5 |
↓
|
|