|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.11 |
4.05 |
-9.13 |
1 |
5 |
0 |
55 |
302.374 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.42 |
10.05 |
-26.94 |
3 |
10 |
0 |
138 |
613.692 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.42 |
11.26 |
-74.85 |
2 |
10 |
-1 |
141 |
612.684 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
23 |
0.31 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
931 |
0.24 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
6.1 |
-19.67 |
2 |
9 |
0 |
116 |
495.557 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.52 |
5.08 |
-54.23 |
1 |
9 |
-1 |
123 |
494.549 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.33 |
7.31 |
-61.73 |
1 |
9 |
-1 |
119 |
494.549 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
23 |
0.31 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
931 |
0.24 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
5.94 |
-24.17 |
2 |
9 |
0 |
116 |
495.557 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.52 |
4.49 |
-54.66 |
1 |
9 |
-1 |
123 |
494.549 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.33 |
7.16 |
-63.52 |
1 |
9 |
-1 |
119 |
494.549 |
6 |
↓
|
|
|
|
Analogs
-
3448206
-
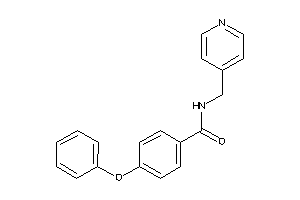
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.29 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
8.46 |
-9.66 |
1 |
4 |
0 |
51 |
318.376 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.44 |
8.92 |
-44.04 |
2 |
4 |
1 |
52 |
319.384 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA10-1-E |
ADAM10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
ADA12-1-E |
ADAM12 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM
|
|
ADA17-1-E |
ADAM17 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
ADAM9-1-E |
ADAM9 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.36 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM
|
|
MMP10-1-E |
Matrix Metalloproteinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.38 |
Binding ≤ 10μM
|
|
MMP11-1-E |
Matrix Metalloproteinase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.38 |
Binding ≤ 10μM
|
|
MMP12-1-E |
Macrophage Metalloelastase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
MMP14-1-E |
Matrix Metalloproteinase 14 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
MMP15-1-E |
Matrix Metalloproteinase 15 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM
|
|
MMP16-1-E |
Matrix Metalloproteinase 16 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
MMP17-1-E |
Matrix Metalloproteinase 17 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
MMP26-1-E |
Matrix Metalloproteinase 26 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.39 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.37 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.55 |
3.16 |
-20.09 |
5 |
8 |
0 |
123 |
388.468 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.55 |
4.38 |
-63.44 |
4 |
8 |
-1 |
126 |
387.46 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA17-1-E |
ADAM17 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.31 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
21 |
0.28 |
Binding ≤ 10μM
|
|
MMP10-1-E |
Matrix Metalloproteinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
190 |
0.25 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.29 |
Binding ≤ 10μM
|
|
MMP14-1-E |
Matrix Metalloproteinase 14 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.22 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
660 |
0.23 |
Binding ≤ 10μM
|
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.31 |
Binding ≤ 10μM
|
|
A4-1-E |
Beta Amyloid A4 Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.25 |
Functional ≤ 10μM
|
|
Z80088-1-O |
CHO (Ovarian Cells) (cluster #1 Of 4), Other |
Other |
200 |
0.25 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
500 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.41 |
12.04 |
-28.44 |
3 |
8 |
0 |
108 |
523.674 |
12 |
↓
|
|
|
|
Analogs
-
11616941
-
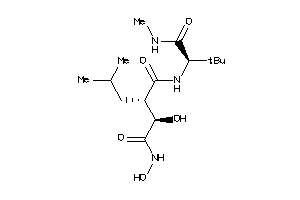
-
11616943
-
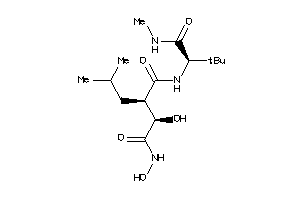
-
27079413
-
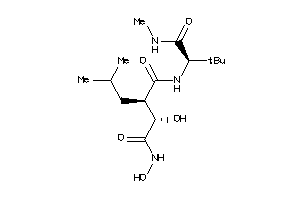
-
43771549
-
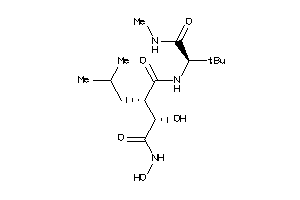
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA17-1-E |
ADAM17 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
MMP14-3-E |
Matrix Metalloproteinase 14 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.43 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.41 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.47 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.49 |
Binding ≤ 10μM
|
|
ADA17-1-E |
ADAM17 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.31 |
Functional ≤ 10μM
|
|
TNFA-2-E |
TNF-alpha (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1001 |
0.37 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
4100 |
0.33 |
Functional ≤ 10μM
|
|
Z80548-2-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #2 Of 5), Other |
Other |
2100 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.38 |
0.45 |
-15.72 |
5 |
8 |
0 |
128 |
331.413 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.38 |
1.66 |
-56.16 |
4 |
8 |
-1 |
131 |
330.405 |
8 |
↓
|
|
|
|
Analogs
-
11616934
-
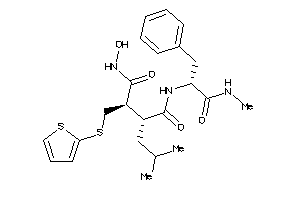
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA17-1-E |
ADAM17 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.34 |
Binding ≤ 10μM
|
|
FCER2-1-E |
Immunoglobulin Epsilon Fc Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.31 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
|
MMP14-1-E |
Matrix Metalloproteinase 14 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
47 |
0.32 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.37 |
Binding ≤ 10μM |
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
20 |
0.34 |
Binding ≤ 10μM |
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.36 |
Binding ≤ 10μM |
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
-4.65 |
-17.84 |
4 |
7 |
0 |
107 |
477.652 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.69 |
-4.09 |
-61.6 |
3 |
7 |
-1 |
113 |
476.644 |
12 |
↓
|
|
|
|
Analogs
-
600699
-
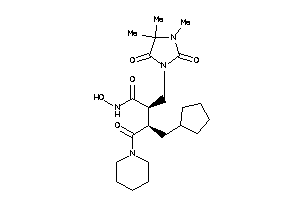
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
527 |
0.28 |
Binding ≤ 10μM |
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
100 |
0.32 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
-1.05 |
-17.22 |
2 |
9 |
0 |
110 |
436.553 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.07 |
-0.49 |
-66.15 |
1 |
9 |
-1 |
113 |
435.545 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
64 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
12.57 |
-7.92 |
0 |
6 |
0 |
70 |
392.436 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4540 |
0.22 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
38 |
0.31 |
Binding ≤ 10μM
|
|
MMP14-1-E |
Matrix Metalloproteinase 14 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
91 |
0.29 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.32 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3025 |
0.23 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
8.67 |
-62.63 |
1 |
9 |
-1 |
125 |
509.626 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8100 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
10.19 |
-32.89 |
0 |
4 |
0 |
40 |
346.477 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
4.1 |
-41.94 |
0 |
5 |
-1 |
57 |
313.387 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.73 |
4.66 |
-10.92 |
1 |
5 |
0 |
60 |
314.395 |
2 |
↓
|
|
|
|
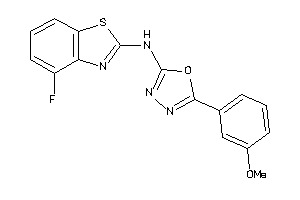
Analogs
-
4137697
-
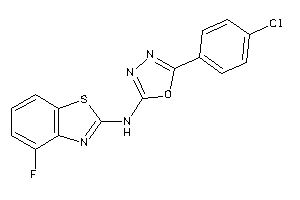
-
4078454
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3500 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.64 |
3.6 |
-18.45 |
1 |
7 |
0 |
82 |
372.381 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4000 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.18 |
7.19 |
-12.67 |
0 |
6 |
0 |
70 |
373.412 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
920 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
7.98 |
-9.9 |
1 |
3 |
0 |
46 |
292.791 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
|
Z102240-1-O |
Cartilage (cluster #1 Of 1), Other |
Other |
1300 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
2.44 |
-58.46 |
0 |
6 |
-1 |
84 |
423.448 |
4 |
↓
|
|
|
|
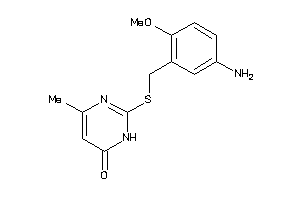
Analogs
-
37011611
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4300 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
5.52 |
-10.08 |
1 |
4 |
0 |
55 |
262.334 |
4 |
↓
|
|
|
|
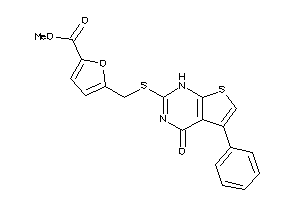
Analogs
-
4289664
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8400 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.65 |
8.09 |
-19.91 |
1 |
6 |
0 |
85 |
404.494 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.11 |
6.87 |
-45.49 |
0 |
6 |
-1 |
88 |
403.486 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1700 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
6.79 |
-8.08 |
1 |
2 |
0 |
33 |
285.393 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.59 |
6.69 |
-37.18 |
2 |
2 |
1 |
34 |
286.401 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.59 |
7.24 |
-32.98 |
2 |
2 |
1 |
34 |
286.401 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
8.27 |
-10.92 |
2 |
6 |
0 |
84 |
410.424 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7700 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
6.09 |
-20.96 |
2 |
8 |
0 |
95 |
432.432 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP12-1-E |
Macrophage Metalloelastase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
320 |
0.31 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
120 |
0.33 |
Binding ≤ 10μM
|
|
MMP14-2-E |
Matrix Metalloproteinase 14 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.29 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.35 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
351 |
0.31 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1300 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.20 |
4.42 |
-55.83 |
2 |
9 |
-1 |
138 |
419.435 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.30 |
6.19 |
-61.75 |
1 |
9 |
-1 |
129 |
431.446 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
9.5 |
-53.18 |
1 |
9 |
-1 |
126 |
451.482 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aR,8aR)-1,4-dioxido-2-(4-phenoxyphenyl)-4a,5,6,7,8,8a-hexahydroquinoxaline-1,4-diium
(4aR,8aR)-1,4-dioxido-2-(4-pheno…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6800 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
9.75 |
-13.53 |
0 |
5 |
0 |
67 |
336.391 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aR,8aS)-1,4-dioxido-2-(4-phenoxyphenyl)-4a,5,6,7,8,8a-hexahydroquinoxaline-1,4-diium
(4aR,8aS)-1,4-dioxido-2-(4-pheno…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6800 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.58 |
9.95 |
-14.28 |
0 |
5 |
0 |
67 |
336.391 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1210 |
0.27 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.38 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
44 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.10 |
5.69 |
-52.56 |
3 |
8 |
1 |
106 |
449.549 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.10 |
6.92 |
-70.06 |
2 |
8 |
0 |
109 |
448.541 |
9 |
↓
|
|