|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BRS3-1-E |
Bombesin Receptor Subtype-3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
GRPR-1-E |
Gastrin Releasing Peptide Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6400 |
0.29 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
1200 |
0.33 |
Binding ≤ 10μM |
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.29 |
Binding ≤ 10μM
|
|
NR1I2-1-E |
Pregnane X Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.31 |
Binding ≤ 10μM |
|
BRS3-1-E |
Bombesin Receptor Subtype-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
99 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.12 |
12.17 |
-37.44 |
2 |
3 |
1 |
43 |
334.487 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.12 |
11.72 |
-9.81 |
1 |
3 |
0 |
42 |
333.479 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.12 |
12.38 |
-9.54 |
1 |
3 |
0 |
42 |
333.479 |
7 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRPR-1-E |
Gastrin Releasing Peptide Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
16 |
0.25 |
Binding ≤ 10μM
|
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.29 |
Binding ≤ 10μM
|
|
GRPR-1-E |
Gastrin Releasing Peptide Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.29 |
Functional ≤ 10μM
|
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.54 |
11.92 |
-20.8 |
4 |
11 |
0 |
154 |
584.677 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.54 |
12.12 |
-43.06 |
5 |
11 |
1 |
155 |
585.685 |
10 |
↓
|
|
|
|
Analogs
-
5190705
-
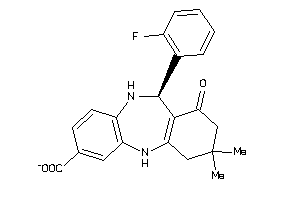
-
5190833
-
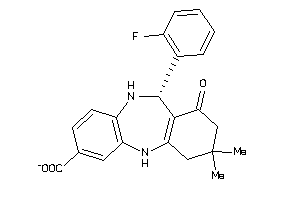
-
720658
-
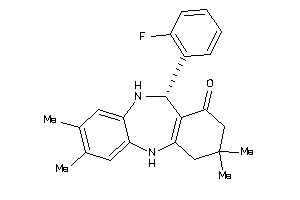
Draw
Identity
99%
90%
80%
70%
Popular Name:
11-(2-fluorophenyl)-3,3,8-trimethyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one
11-(2-fluorophenyl)-3,3,8-trimet…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
0.41 |
-11.32 |
2 |
3 |
0 |
41 |
350.437 |
1 |
↓
|
|
|
|
Analogs
-
5190705
-

-
5190833
-

-
720658
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
11-(2-fluorophenyl)-3,3,8-trimethyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one
11-(2-fluorophenyl)-3,3,8-trimet…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
0.42 |
-11.26 |
2 |
3 |
0 |
41 |
350.437 |
1 |
↓
|
|
|
|
Analogs
-
4406262
-
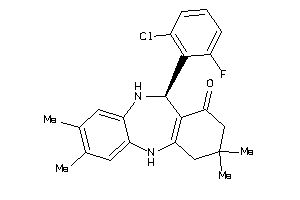
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.34 |
Binding ≤ 10μM
|
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.51 |
-0.05 |
-8.73 |
2 |
3 |
0 |
41 |
384.882 |
1 |
↓
|
|
|
|
Analogs
-
4406262
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.34 |
Binding ≤ 10μM
|
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.51 |
0.02 |
-8.54 |
2 |
3 |
0 |
41 |
384.882 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6900 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.77 |
-0.23 |
-8.68 |
2 |
3 |
0 |
41 |
332.447 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6900 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.77 |
-0.23 |
-8.36 |
2 |
3 |
0 |
41 |
332.447 |
1 |
↓
|
|
|
|
Analogs
-
672448
-
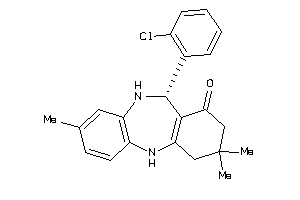
-
5364102
-
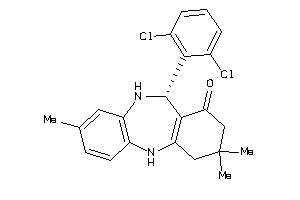
-
5364106
-
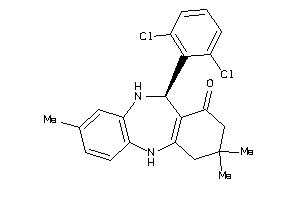
-
18190063
-
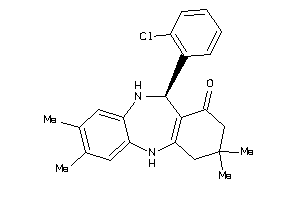
-
18190065
-
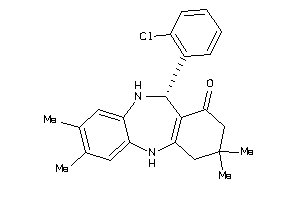
Draw
Identity
99%
90%
80%
70%
Popular Name:
11-(2-chlorophenyl)-3,3,8-trimethyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one
11-(2-chlorophenyl)-3,3,8-trimet…
Find On:
PubMed —
Wikipedia —
Google
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
280 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.40 |
-0.34 |
-8.83 |
2 |
3 |
0 |
41 |
366.892 |
1 |
↓
|
|
|
|
Analogs
-
5364102
-

-
5364106
-

-
18190063
-

-
18190065
-

-
672446
-
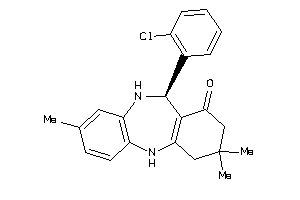
Draw
Identity
99%
90%
80%
70%
Popular Name:
11-(2-chlorophenyl)-3,3,8-trimethyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one
11-(2-chlorophenyl)-3,3,8-trimet…
Find On:
PubMed —
Wikipedia —
Google
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
280 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.40 |
-0.35 |
-8.86 |
2 |
3 |
0 |
41 |
366.892 |
1 |
↓
|
|
|
|
Analogs
-
5190760
-
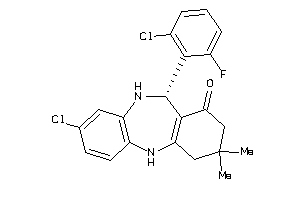
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.74 |
-0.52 |
-8.87 |
2 |
3 |
0 |
41 |
405.3 |
1 |
↓
|
|
|
|
Analogs
-
5190626
-
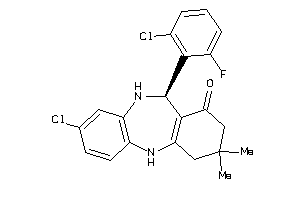
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMBR-1-E |
Neuromedin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.74 |
-0.52 |
-8.86 |
2 |
3 |
0 |
41 |
405.3 |
1 |
↓
|
|
|
|
|