|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
2400 |
0.79 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
1600 |
0.81 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
2.75 |
-28.71 |
3 |
2 |
1 |
40 |
141.238 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
2400 |
0.79 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
1600 |
0.81 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
2.75 |
-28.69 |
3 |
2 |
1 |
40 |
141.238 |
1 |
↓
|
|
|
|
Analogs
-
26377093
-
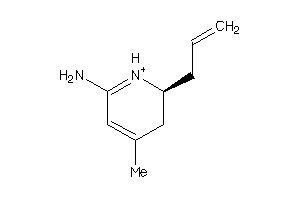
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
50 |
0.93 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
360 |
0.82 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
3.98 |
-26.41 |
3 |
2 |
1 |
40 |
151.233 |
2 |
↓
|
|
|
|
Analogs
-
13476792
-
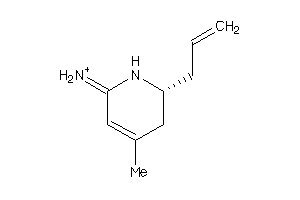
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
50 |
0.93 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
360 |
0.82 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
3.94 |
-26.42 |
3 |
2 |
1 |
40 |
151.233 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
200 |
1.17 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
230 |
1.16 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.85 |
1.92 |
-28.02 |
3 |
2 |
1 |
40 |
111.168 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
610 |
0.97 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
930 |
0.94 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
2.37 |
-28.92 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
610 |
0.97 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
930 |
0.94 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
2.54 |
-28.91 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
610 |
0.97 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
930 |
0.94 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
2.43 |
-28.81 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
610 |
0.97 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
930 |
0.94 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
2.33 |
-28.92 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
170 |
1.05 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
170 |
1.05 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1600 |
0.90 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
2.84 |
-28.83 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
170 |
1.05 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
170 |
1.05 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1600 |
0.90 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
2.73 |
-28.82 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
170 |
1.05 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
170 |
1.05 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1600 |
0.90 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
2.84 |
-28.8 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
170 |
1.05 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
170 |
1.05 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1600 |
0.90 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
2.72 |
-28.86 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
390 |
0.90 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
300 |
0.91 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3500 |
0.76 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
3.6 |
-29.3 |
3 |
2 |
1 |
40 |
141.238 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
390 |
0.90 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
300 |
0.91 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3500 |
0.76 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
3.52 |
-29.34 |
3 |
2 |
1 |
40 |
141.238 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
390 |
0.90 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
300 |
0.91 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3500 |
0.76 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
3.6 |
-29.3 |
3 |
2 |
1 |
40 |
141.238 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
390 |
0.90 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
300 |
0.91 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3500 |
0.76 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
3.52 |
-29.34 |
3 |
2 |
1 |
40 |
141.238 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4R,4aR,7aR)-4-methyl-4,4a,5,6,7,7a-hexahydro-3H-cyclopenta[e]pyridin-2-amine
(4R,4aR,7aR)-4-methyl-4,4a,5,6,7…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
31 |
0.96 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
9 |
1.02 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
470 |
0.81 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
3.74 |
-29.29 |
3 |
2 |
1 |
40 |
153.249 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
400 |
1.12 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
500 |
1.10 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
600 |
1.09 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.89 |
2.17 |
-28.15 |
3 |
2 |
1 |
40 |
113.184 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
400 |
1.12 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
500 |
1.10 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
600 |
1.09 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.89 |
2.18 |
-28.17 |
3 |
2 |
1 |
40 |
113.184 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
700 |
1.08 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
6000 |
0.91 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3000 |
0.97 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
2.09 |
-28.21 |
3 |
2 |
1 |
40 |
113.184 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
700 |
1.08 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
6000 |
0.91 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3000 |
0.97 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
2.1 |
-28.22 |
3 |
2 |
1 |
40 |
113.184 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
4500 |
0.94 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
5100 |
0.93 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
2.07 |
-29.2 |
3 |
2 |
1 |
40 |
113.184 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
4500 |
0.94 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
5100 |
0.93 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
2.07 |
-29.17 |
3 |
2 |
1 |
40 |
113.184 |
0 |
↓
|
|
|
|
Analogs
-
33685532
-
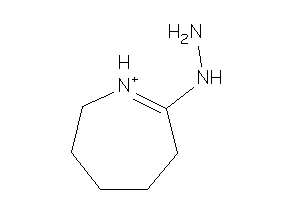
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
3500 |
0.95 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
2100 |
0.99 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.06 |
1.91 |
-29.08 |
3 |
2 |
1 |
40 |
113.184 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
140 |
1.07 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
160 |
1.06 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6300 |
0.81 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.36 |
2.59 |
-29 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
140 |
1.07 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
160 |
1.06 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6300 |
0.81 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.36 |
2.73 |
-29.02 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
140 |
1.07 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
160 |
1.06 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6300 |
0.81 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.36 |
2.72 |
-28.98 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
140 |
1.07 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
160 |
1.06 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6300 |
0.81 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.36 |
2.59 |
-29.03 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
3500 |
0.85 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
2.56 |
-29.93 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
3500 |
0.85 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
2.53 |
-29.92 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
-
34559376
-

-
34559377
-
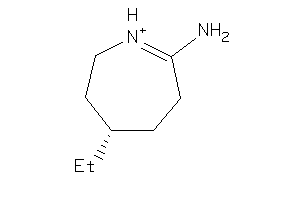
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
5800 |
0.81 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.07 |
2.5 |
-29.64 |
3 |
2 |
1 |
40 |
127.211 |
0 |
↓
|
|
|
|
Analogs
-
34559376
-

-
34559377
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
5800 |
0.81 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.07 |
2.49 |
-29.68 |
3 |
2 |
1 |
40 |
127.211 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
200 |
1.17 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
37 |
1.30 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
220 |
1.17 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
1.94 |
-27.24 |
3 |
2 |
1 |
40 |
113.184 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
200 |
1.17 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
37 |
1.30 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
220 |
1.17 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
2.1 |
-29.22 |
3 |
2 |
1 |
40 |
113.184 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
1600 |
0.90 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
2600 |
0.87 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.48 |
2.77 |
-29.56 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
1600 |
0.90 |
Binding ≤ 10μM
|
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
2600 |
0.87 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.48 |
2.76 |
-29.57 |
3 |
2 |
1 |
40 |
127.211 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS2-1-E |
Nitric Oxide Synthase, Inducible (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
700 |
1.08 |
Binding ≤ 10μM
|
|
NOS3-7-E |
Nitric Oxide Synthase, Endothelial (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
440 |
1.11 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.84 |
2.07 |
-28.28 |
3 |
2 |
1 |
40 |
111.168 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.02 |
1.37 |
-41.14 |
4 |
2 |
1 |
40 |
111.168 |
0 |
↓
|
|