|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
850 |
0.40 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4650 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
7.69 |
-7.26 |
1 |
4 |
0 |
51 |
278.311 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
260 |
0.33 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
275 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.76 |
10.28 |
-9.1 |
1 |
5 |
0 |
60 |
370.408 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.34 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.08 |
11 |
-8.29 |
1 |
5 |
0 |
60 |
384.435 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.39 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
7.97 |
-8 |
1 |
5 |
0 |
60 |
322.364 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
840 |
0.37 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
840 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.49 |
6.98 |
-8.21 |
1 |
5 |
0 |
60 |
308.337 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.30 |
21.53 |
-23.54 |
0 |
7 |
0 |
96 |
614.779 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
6.74 |
-7.68 |
1 |
4 |
0 |
50 |
373.293 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.48 |
7.2 |
-40.27 |
0 |
4 |
-1 |
53 |
372.285 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
93 |
0.25 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.29 |
Binding ≤ 10μM
|
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.25 |
Functional ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
240 |
0.24 |
Functional ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1312 |
0.21 |
Functional ≤ 10μM
|
|
PPARD-1-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
680 |
0.22 |
Functional ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
680 |
0.22 |
Functional ≤ 10μM
|
|
Z80169-1-O |
Huh-7 (Hepatocellular Carcinoma) (cluster #1 Of 1), Other |
Other |
45 |
0.26 |
Functional ≤ 10μM
|
|
Z80178-3-O |
J774.A1 (Macrophage Cells) (cluster #3 Of 3), Other |
Other |
41 |
0.27 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
84 |
0.25 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
223 |
0.24 |
Functional ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
41 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.81 |
17.18 |
-54.11 |
0 |
4 |
-1 |
62 |
526.534 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.37 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1320 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.02 |
11.6 |
-7.68 |
1 |
4 |
0 |
51 |
354.409 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
560 |
0.40 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
890 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
8.68 |
-7.36 |
1 |
4 |
0 |
51 |
292.338 |
4 |
↓
|
|
|
|
Analogs
-
4177478
-
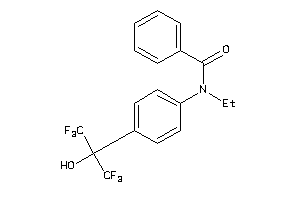
-
828594
-
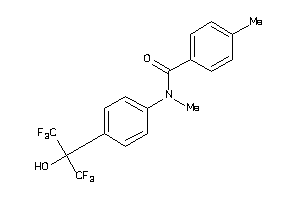
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.32 |
3.22 |
-13.15 |
1 |
3 |
0 |
40 |
377.284 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]benzenesulfonamide
N-[4-(1,1,1,3,3,3-hexafluoro-2-h…
Find On:
PubMed —
Wikipedia —
Google
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
3.66 |
-10.44 |
2 |
4 |
0 |
66 |
399.312 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.95 |
4.28 |
-102.14 |
0 |
4 |
-2 |
71 |
397.296 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.95 |
3.76 |
-41.73 |
1 |
4 |
-1 |
68 |
398.304 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.67 |
12.13 |
-12.23 |
0 |
5 |
0 |
51 |
492.906 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.67 |
11.48 |
-11.42 |
0 |
5 |
0 |
51 |
492.906 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.74 |
12.88 |
-8.73 |
0 |
2 |
0 |
18 |
422.784 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.12 |
8.75 |
-8.02 |
2 |
3 |
0 |
44 |
490.271 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
7.12 |
9.99 |
-87.23 |
0 |
3 |
-2 |
49 |
488.255 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
7.12 |
9.51 |
-40.68 |
1 |
3 |
-1 |
47 |
489.263 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-2-E |
LXR-beta (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
175 |
0.31 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
315 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.08 |
3.82 |
-6.67 |
1 |
2 |
0 |
23 |
439.399 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.76 |
17.52 |
-57.16 |
0 |
4 |
-1 |
53 |
581.054 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.12 |
6.6 |
-15.78 |
1 |
4 |
0 |
58 |
481.336 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.12 |
7.07 |
-43.66 |
0 |
4 |
-1 |
60 |
480.328 |
8 |
↓
|
|