|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.69 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.69 |
Binding ≤ 10μM
|
|
Q8CFB8-1-E |
ADP-ribosyltransferase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
0.61 |
-11.07 |
2 |
4 |
0 |
66 |
176.175 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
0.80 |
0.77 |
-41.66 |
1 |
4 |
-1 |
69 |
175.167 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.80 |
1.37 |
-48.29 |
1 |
4 |
-1 |
69 |
175.167 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
870 |
0.47 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
870 |
0.47 |
Binding ≤ 10μM
|
|
Q8CFB8-1-E |
ADP-ribosyltransferase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
870 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
7.46 |
-9.08 |
1 |
3 |
0 |
46 |
236.274 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.96 |
5.61 |
-52.49 |
0 |
3 |
-1 |
49 |
235.266 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
850 |
0.39 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
850 |
0.39 |
Binding ≤ 10μM
|
|
Q8CFB8-1-E |
ADP-ribosyltransferase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
850 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
6.66 |
-12.58 |
1 |
7 |
0 |
101 |
297.27 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
520 |
0.46 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.46 |
Binding ≤ 10μM
|
|
Q8CFB8-1-E |
ADP-ribosyltransferase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
1.97 |
-45.12 |
3 |
5 |
-1 |
95 |
252.253 |
1 |
↓
|
|
|
|
Analogs
-
39343284
-
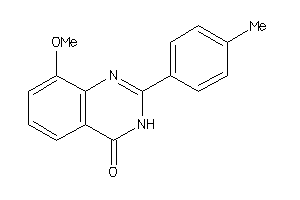
-
5335585
-
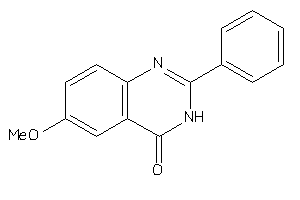
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.40 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.40 |
Binding ≤ 10μM
|
|
Q8CFB8-1-E |
ADP-ribosyltransferase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.11 |
5.99 |
-10.94 |
1 |
4 |
0 |
55 |
252.273 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
290 |
0.48 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.48 |
Binding ≤ 10μM
|
|
Q8CFB8-1-E |
ADP-ribosyltransferase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
1.27 |
-42.74 |
2 |
5 |
-1 |
89 |
253.237 |
1 |
↓
|
|
|
|
Analogs
-
39123625
-
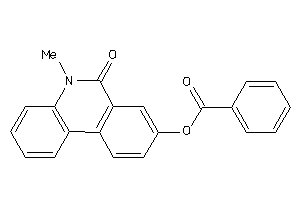
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.38 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.32 |
7.92 |
-13.07 |
1 |
4 |
0 |
59 |
265.268 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
230 |
0.44 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
230 |
0.44 |
Binding ≤ 10μM
|
|
Q8CFB8-1-E |
ADP-ribosyltransferase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
230 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.79 |
4.56 |
-12.42 |
2 |
7 |
0 |
112 |
283.243 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.25 |
3.49 |
-105.16 |
0 |
7 |
-2 |
118 |
281.227 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.79 |
5.32 |
-44.39 |
1 |
7 |
-1 |
115 |
282.235 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1340 |
0.39 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1340 |
0.39 |
Binding ≤ 10μM
|
|
Q8CFB8-1-E |
ADP-ribosyltransferase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1340 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
6.38 |
-12.82 |
1 |
5 |
0 |
79 |
277.283 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.38 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.38 |
Binding ≤ 10μM
|
|
Q8CFB8-1-E |
ADP-ribosyltransferase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.17 |
5.3 |
-11.45 |
1 |
5 |
0 |
64 |
282.299 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1060 |
0.46 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1060 |
0.46 |
Binding ≤ 10μM
|
|
Q8CFB8-1-E |
ADP-ribosyltransferase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1060 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
4.1 |
-43.3 |
1 |
4 |
-1 |
69 |
237.238 |
1 |
↓
|
|
|
|
Analogs
-
34580013
-
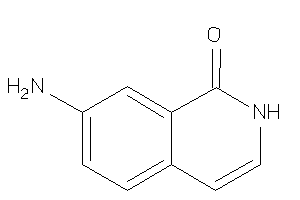
-
34580014
-
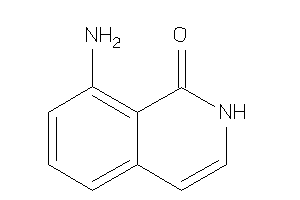
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
240 |
0.77 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1050 |
0.70 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.93 |
1.69 |
-11.1 |
3 |
3 |
0 |
59 |
160.176 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.55 |
5.17 |
-52.77 |
5 |
5 |
1 |
88 |
325.367 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.47 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.55 |
5.17 |
-52.45 |
5 |
5 |
1 |
88 |
325.367 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.54 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.59 |
5.27 |
-57.19 |
4 |
5 |
1 |
76 |
287.387 |
4 |
↓
|
|
|
|
Analogs
-
35384958
-
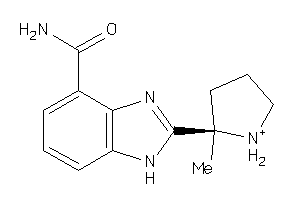
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.65 |
Binding ≤ 10μM
|
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.65 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.68 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.90 |
1.66 |
-37.4 |
5 |
5 |
1 |
88 |
245.306 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
0.90 |
1.64 |
-38.5 |
5 |
5 |
1 |
88 |
245.306 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.90 |
0.43 |
-12.69 |
4 |
5 |
0 |
84 |
244.298 |
2 |
↓
|
|
|
|
Analogs
-
36470974
-
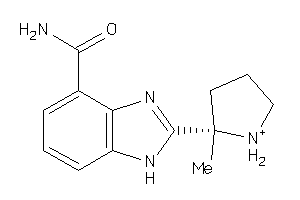
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.65 |
Binding ≤ 10μM
|
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.65 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.65 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.90 |
1.54 |
-39.23 |
5 |
5 |
1 |
88 |
245.306 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.90 |
0.28 |
-16.07 |
4 |
5 |
0 |
84 |
244.298 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.60 |
4.74 |
-96.74 |
7 |
8 |
2 |
122 |
420.517 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Binding ≤ 10μM
|
|
PARP2-1-E |
Poly [ADP-ribose] Polymerase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
5.86 |
-20.8 |
3 |
5 |
0 |
85 |
314.348 |
3 |
↓
|
|