|
|
Analogs
-
60292553
-
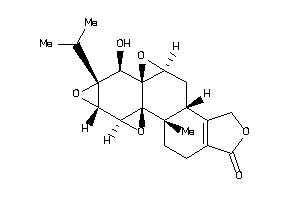
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH2-7-E |
Cyclooxygenase-2 (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
40 |
0.40 |
Binding ≤ 10μM
|
|
Z80125-2-O |
DU-145 (Prostate Carcinoma) (cluster #2 Of 9), Other |
Other |
24 |
0.41 |
Functional ≤ 10μM
|
|
Z80166-2-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #2 Of 12), Other |
Other |
2 |
0.47 |
Functional ≤ 10μM
|
|
Z80186-4-O |
K562 (Erythroleukemia Cells) (cluster #4 Of 11), Other |
Other |
50 |
0.39 |
Functional ≤ 10μM
|
|
Z80224-2-O |
MCF7 (Breast Carcinoma Cells) (cluster #2 Of 14), Other |
Other |
19 |
0.42 |
Functional ≤ 10μM
|
|
Z80244-3-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #3 Of 7), Other |
Other |
10 |
0.43 |
Functional ≤ 10μM
|
|
Z80390-2-O |
PC-3 (Prostate Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
43 |
0.40 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z80559-2-O |
U251 (cluster #2 Of 3), Other |
Other |
49 |
0.39 |
Functional ≤ 10μM
|
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
59 |
0.39 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
10 |
0.43 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
43 |
0.40 |
Functional ≤ 10μM
|
|
Z81247-3-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #3 Of 9), Other |
Other |
47 |
0.39 |
Functional ≤ 10μM
|
|
Z81252-4-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #4 Of 11), Other |
Other |
24 |
0.41 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
29 |
0.41 |
Functional ≤ 10μM
|
|
Z80253-2-O |
MEF (Embryonic Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
9200 |
0.27 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.57 |
-5.07 |
-17.26 |
1 |
6 |
0 |
84 |
360.406 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH2-7-E |
Cyclooxygenase-2 (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
3.15 |
8.88 |
-8.32 |
1 |
5 |
0 |
94 |
361.458 |
7 |
↓
|
|
|
|
Analogs
-
57313
-
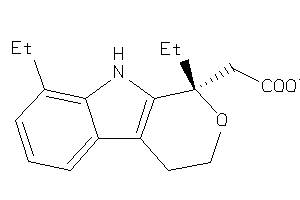
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH2-7-E |
Cyclooxygenase-2 (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
2000 |
0.38 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.51 |
Functional ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
23 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.64 |
9.1 |
-51.62 |
1 |
4 |
-1 |
65 |
286.351 |
4 |
↓
|
|
|
|
Analogs
-
3642
-
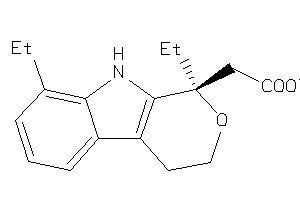
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH2-7-E |
Cyclooxygenase-2 (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
2000 |
0.38 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.51 |
Functional ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
23 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.64 |
9.02 |
-46.89 |
1 |
4 |
-1 |
65 |
286.351 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH2-7-E |
Cyclooxygenase-2 (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
140 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
-1.29 |
-17.26 |
0 |
5 |
0 |
69 |
336.409 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3aS,6S,7R,8S,8aR)-8-hydroxy-6,8-dimethyl-3-methylene-spiro[4,5,6,8a-tetrahydro-3aH-cyclohepta[b]fur
(3aS,6S,7R,8S,8aR)-8-hydroxy-6,8…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH2-7-E |
Cyclooxygenase-2 (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
4600 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.83 |
5.66 |
-21.2 |
1 |
5 |
0 |
73 |
280.32 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH2-7-E |
Cyclooxygenase-2 (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
40 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.56 |
16.12 |
-10.15 |
0 |
5 |
0 |
58 |
455.982 |
11 |
↓
|
|
|
|
Analogs
-
33838895
-
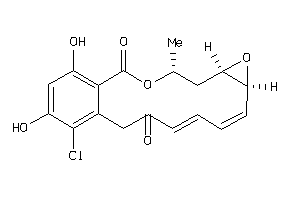
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CBR1-1-E |
Carbonyl Reductase [NADPH] 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.35 |
Binding ≤ 10μM
|
|
HS90A-1-E |
Heat Shock Protein HSP 90-alpha (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
530 |
0.35 |
Binding ≤ 10μM |
|
HS90B-1-E |
Heat Shock Protein HSP 90-beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.43 |
Binding ≤ 10μM
|
|
PGH2-7-E |
Cyclooxygenase-2 (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
27 |
0.42 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
180 |
0.38 |
Binding ≤ 10μM
|
|
HSC82-1-F |
Heat Shock Protein HSP 90 (HSC82) (cluster #1 Of 1), Fungal |
Fungi |
200 |
0.38 |
Binding ≤ 10μM
|
|
HSP82-1-F |
Heat Shock Protein HSP 90 (HSP82) (cluster #1 Of 1), Fungal |
Fungi |
19 |
0.43 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7200 |
0.29 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
200 |
0.38 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
89 |
0.39 |
Functional ≤ 10μM
|
|
Z80339-1-O |
NRK (cluster #1 Of 1), Other |
Other |
290 |
0.37 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
38 |
0.42 |
Functional ≤ 10μM
|
|
Z80622-1-O |
3Y1 Cell Line (cluster #1 Of 1), Other |
Other |
700 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
3.67 |
-13.53 |
2 |
6 |
0 |
96 |
364.781 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.70 |
5.33 |
-120.39 |
0 |
6 |
-2 |
102 |
362.765 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.70 |
4.33 |
-46.69 |
1 |
6 |
-1 |
99 |
363.773 |
0 |
↓
|
|