|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PNPH-1-E |
Purine-nucleoside Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.73 |
2.66 |
-27.39 |
4 |
6 |
0 |
100 |
247.283 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PNPH-1-E |
Purine-nucleoside Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.04 |
2.85 |
-27.63 |
4 |
6 |
0 |
100 |
247.283 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PNPH-1-E |
Purine-nucleoside Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.14 |
4.68 |
-18.74 |
4 |
6 |
0 |
100 |
241.254 |
2 |
↓
|
|
|
|
Analogs
-
38804439
-
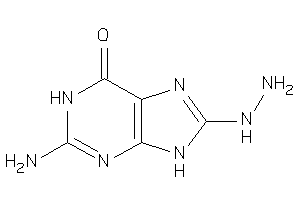
Draw
Identity
99%
90%
80%
70%
Vendors
And 106 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PNPH-1-E |
Purine-nucleoside Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.67 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.93 |
1.36 |
-17.89 |
4 |
6 |
0 |
100 |
151.129 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PNPH-1-E |
Purine-nucleoside Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.68 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.21 |
-0.73 |
-56.39 |
3 |
6 |
0 |
89 |
234.259 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.67 |
-2.37 |
-19.04 |
3 |
6 |
0 |
85 |
234.259 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.21 |
-3.5 |
-51.64 |
2 |
6 |
-1 |
88 |
233.251 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PNPH-1-E |
Purine-nucleoside Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
57 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.12 |
1.38 |
-30.04 |
4 |
7 |
0 |
113 |
228.215 |
1 |
↓
|
|
|
|
Analogs
-
17013444
-
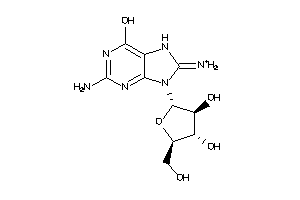
-
4916515
-
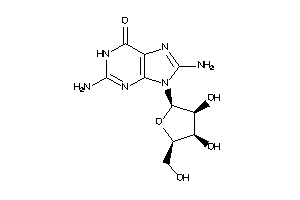
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PNPH-1-E |
Purine-nucleoside Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.34 |
Binding ≤ 10μM
|
|
PNPH-1-E |
Purine Nucleoside Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1330 |
0.39 |
Functional ≤ 10μM
|
|
Z80285-1-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
3250 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.21 |
-5.54 |
-23.28 |
8 |
11 |
0 |
186 |
298.259 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PNPH-1-E |
Purine-nucleoside Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9900 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.59 |
-0.16 |
-34.75 |
7 |
6 |
1 |
115 |
166.164 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.59 |
-5.97 |
-33.91 |
7 |
6 |
1 |
114 |
166.164 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.59 |
-0.62 |
-19.19 |
6 |
6 |
0 |
114 |
165.156 |
0 |
↓
|
|
|
|
Analogs
-
38804439
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PNPH-1-E |
Purine-nucleoside Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.82 |
Binding ≤ 10μM
|
|
PNPH-1-E |
Purine Nucleoside Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1550 |
0.68 |
Functional ≤ 10μM
|
|
Z80285-3-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #3 Of 4), Other |
Other |
2350 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.12 |
0.9 |
-17.55 |
6 |
7 |
0 |
126 |
166.144 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
-1.12 |
0.81 |
-14.79 |
6 |
7 |
0 |
126 |
166.144 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PNPH-1-E |
Purine-nucleoside Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.49 |
Binding ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
770 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.54 |
6.4 |
-24.53 |
5 |
7 |
0 |
116 |
256.269 |
2 |
↓
|
|