|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLAP-1-E |
Phospholipase A-2-activating Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.32 |
Binding ≤ 10μM
|
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
3.06 |
-14.56 |
2 |
8 |
0 |
110 |
358.379 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.75 |
3.99 |
-46.29 |
1 |
8 |
-1 |
113 |
357.371 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 40 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
4.23 |
-43.29 |
1 |
5 |
-1 |
78 |
215.188 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
6.36 |
-6.6 |
1 |
4 |
0 |
55 |
271.103 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
44 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
-6.91 |
-10.68 |
4 |
5 |
0 |
83 |
271.107 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 47 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
5.79 |
-44.18 |
1 |
4 |
-1 |
69 |
221.623 |
2 |
↓
|
|
|
|
Analogs
-
6706918
-
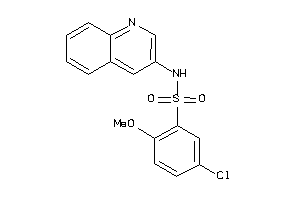
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
540 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
3.8 |
-12.6 |
1 |
5 |
0 |
68 |
312.778 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1550 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
2.54 |
-10.44 |
1 |
6 |
0 |
78 |
373.228 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.41 |
2.38 |
-47.05 |
0 |
6 |
-1 |
80 |
372.22 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1130 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
3.95 |
-12.73 |
1 |
5 |
0 |
68 |
292.36 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.44 |
4.01 |
-54.37 |
0 |
5 |
-1 |
70 |
291.352 |
5 |
↓
|
|
|
|
Analogs
-
6706918
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
590 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.90 |
3.86 |
-56.17 |
0 |
6 |
-1 |
80 |
343.384 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.90 |
3.98 |
-12.19 |
1 |
6 |
0 |
78 |
344.392 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.90 |
4.29 |
-28.21 |
1 |
6 |
0 |
81 |
344.392 |
5 |
↓
|
|
|
|
Analogs
-
37054975
-
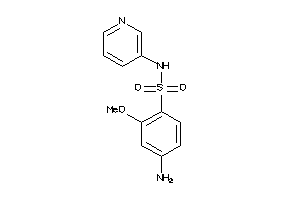
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1060 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
1.86 |
-12.45 |
1 |
6 |
0 |
78 |
294.332 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.67 |
1.71 |
-54.69 |
0 |
6 |
-1 |
80 |
293.324 |
5 |
↓
|
|
|
|
Analogs
-
37054975
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1290 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
2.64 |
-11.08 |
1 |
5 |
0 |
68 |
282.296 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.78 |
2.49 |
-50.35 |
0 |
5 |
-1 |
70 |
281.288 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1490 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
2.61 |
-11.49 |
1 |
6 |
0 |
78 |
308.359 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
690 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
4.39 |
-12.62 |
1 |
5 |
0 |
68 |
326.805 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
3.78 |
-11.3 |
1 |
5 |
0 |
68 |
292.36 |
4 |
↓
|
|
|
|
Analogs
-
6706918
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
5.2 |
-51.36 |
0 |
5 |
-1 |
70 |
392.254 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.65 |
5.63 |
-24.94 |
1 |
5 |
0 |
72 |
393.262 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.65 |
5.31 |
-10.24 |
1 |
5 |
0 |
68 |
393.262 |
4 |
↓
|
|
|
|
Analogs
-
6706918
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
650 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
5.24 |
-55.29 |
0 |
5 |
-1 |
70 |
327.385 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.29 |
5.35 |
-11.12 |
1 |
5 |
0 |
68 |
328.393 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.29 |
5.67 |
-26.98 |
1 |
5 |
0 |
72 |
328.393 |
4 |
↓
|
|
|
|
Analogs
-
37054975
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1850 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
3.23 |
-11.21 |
1 |
5 |
0 |
68 |
278.333 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.06 |
3.09 |
-53.8 |
0 |
5 |
-1 |
70 |
277.325 |
4 |
↓
|
|
|
|
Analogs
-
37054975
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3160 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.58 |
3.13 |
-42.73 |
0 |
8 |
-1 |
116 |
308.295 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.58 |
3.27 |
-10.95 |
1 |
8 |
0 |
114 |
309.303 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.58 |
3.62 |
-31.75 |
1 |
8 |
0 |
117 |
309.303 |
5 |
↓
|
|
|
|
Analogs
-
6706918
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
3.66 |
-10.52 |
1 |
5 |
0 |
68 |
312.778 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1130 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
2.42 |
-10.51 |
1 |
6 |
0 |
78 |
328.777 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.28 |
2.27 |
-47.11 |
0 |
6 |
-1 |
80 |
327.769 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLAP-1-E |
Phospholipase A-2-activating Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.39 |
Binding ≤ 10μM
|
|
PPBN-1-E |
Alkaline Phosphatase Placental-like (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.44 |
Binding ≤ 10μM
|
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8600 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
3.05 |
-15.56 |
2 |
5 |
0 |
83 |
290.344 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.33 |
3.99 |
-47.82 |
1 |
5 |
-1 |
86 |
289.336 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLAP-1-E |
Phospholipase A-2-activating Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.33 |
Binding ≤ 10μM
|
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4800 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
3.44 |
-15.65 |
2 |
6 |
0 |
103 |
344.392 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.84 |
4.38 |
-47.18 |
1 |
6 |
-1 |
106 |
343.384 |
6 |
↓
|
|
|
|
Analogs
-
1492564
-
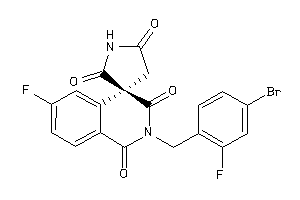
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AK1A1-1-E |
Aldehyde Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.37 |
Binding ≤ 10μM
|
|
AK1BA-1-E |
Aldo-keto Reductase Family 1 Member B10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.31 |
Binding ≤ 10μM
|
|
ALDR-2-E |
Aldose Reductase (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
14 |
0.39 |
Binding ≤ 10μM
|
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.08 |
-0.4 |
-10.22 |
1 |
6 |
0 |
83 |
449.207 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
10.02 |
-52.28 |
0 |
4 |
-1 |
53 |
356.345 |
6 |
↓
|
|