|
|
Analogs
-
12504292
-
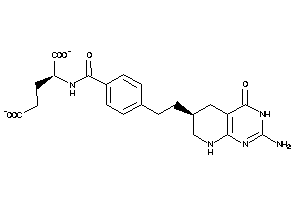
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.30 |
Binding ≤ 10μM |
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.27 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.33 |
Binding ≤ 10μM
|
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.30 |
Functional ≤ 10μM |
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
59 |
0.32 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
8 |
0.35 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
16 |
0.34 |
Functional ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
18 |
0.34 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
1500 |
0.25 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
1500 |
0.25 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
38 |
0.32 |
Functional ≤ 10μM |
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
31 |
0.33 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
31 |
0.33 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.12 |
8.18 |
-127.86 |
5 |
11 |
-2 |
193 |
441.444 |
9 |
↓
|
|
Ref
Reference (pH 7)
|
-1.12 |
7.51 |
-139.85 |
5 |
11 |
-2 |
193 |
441.444 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.12 |
5.53 |
-83.64 |
6 |
11 |
-1 |
190 |
442.452 |
9 |
↓
|
|
|
|
Analogs
-
21289577
-
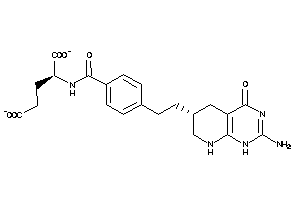
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
60 |
0.32 |
Binding ≤ 10μM |
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.30 |
Binding ≤ 10μM |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.33 |
Binding ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
59 |
0.32 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
15 |
0.34 |
Functional ≤ 10μM |
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
16 |
0.34 |
Functional ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
18 |
0.34 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
1500 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.12 |
5.35 |
-138.84 |
5 |
11 |
-2 |
193 |
441.444 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.12 |
4.23 |
-86.52 |
6 |
11 |
-1 |
190 |
442.452 |
9 |
↓
|
|
|
|
Analogs
-
1851132
-
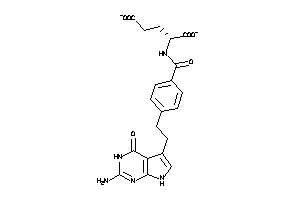
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.29 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
380 |
0.29 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
340 |
0.29 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
2300 |
0.25 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
1100 |
0.27 |
Binding ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
16 |
0.35 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
22 |
0.35 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
894 |
0.27 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
68 |
0.32 |
Functional ≤ 10μM |
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
894 |
0.27 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.53 |
8.7 |
-125.85 |
5 |
11 |
-2 |
197 |
425.401 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.53 |
6.72 |
-70.62 |
6 |
11 |
-1 |
194 |
426.409 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2060 |
0.27 |
Binding ≤ 10μM
|
|
Z80285-1-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
1600 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.02 |
3.88 |
-138.28 |
7 |
12 |
-2 |
219 |
436.45 |
11 |
↓
|
|
Ref
Reference (pH 7)
|
-2.02 |
4.52 |
-127.06 |
7 |
12 |
-2 |
219 |
436.45 |
11 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.02 |
2.59 |
-68.89 |
8 |
12 |
-1 |
216 |
437.458 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
60 |
0.32 |
Binding ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
43 |
0.32 |
Functional ≤ 10μM |
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
1 |
0.39 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.54 |
9.19 |
-125.01 |
5 |
11 |
-2 |
197 |
459.484 |
11 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.54 |
7.26 |
-68.21 |
6 |
11 |
-1 |
194 |
460.492 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.36 |
-8.07 |
-131.56 |
5 |
11 |
-2 |
193 |
465.513 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
22 |
0.31 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
90 |
0.28 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
95 |
0.28 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
430 |
0.25 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.28 |
Binding ≤ 10μM
|
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.21 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
360 |
0.26 |
Binding ≤ 10μM
|
|
Z50266-1-O |
Enterococcus Faecium (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
750 |
0.24 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
5 |
0.33 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7000 |
0.21 |
Functional ≤ 10μM
|
|
Z80443-1-O |
S180 (Sarcoma Cells) (cluster #1 Of 1), Other |
Other |
1730 |
0.23 |
Functional ≤ 10μM
|
|
Z80830-1-O |
Ehrlich (Ehrlich Ascites Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1820 |
0.23 |
Functional ≤ 10μM
|
|
Z80860-1-O |
GC3/M Cell Line (cluster #1 Of 1), Other |
Other |
2130 |
0.23 |
Functional ≤ 10μM
|
|
Z80862-1-O |
GC3/MTK- Cell Line (cluster #1 Of 1), Other |
Other |
1400 |
0.23 |
Functional ≤ 10μM
|
|
Z80890-1-O |
H350.3 (MTX-resistant Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z81221-1-O |
Lymphoma Cell Line (cluster #1 Of 1), Other |
Other |
560 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.82 |
8.37 |
-132.31 |
4 |
11 |
-2 |
184 |
475.461 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C3SWJ7-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
19 |
0.31 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
22 |
0.31 |
Binding ≤ 10μM
|
|
E2QDN0-1-B |
Thymidylate Synthase (EC 2.1.1.45) (TS) (TSase) (cluster #1 Of 1), Bacterial |
Bacteria |
48 |
0.29 |
Binding ≤ 10μM |
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
90 |
0.28 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
95 |
0.28 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.29 |
Binding ≤ 10μM |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
430 |
0.25 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.28 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.28 |
Binding ≤ 10μM
|
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.21 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
360 |
0.26 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
360 |
0.26 |
Binding ≤ 10μM
|
|
Z100337-1-O |
W1-L2 (cluster #1 Of 1), Other |
Other |
440 |
0.25 |
Functional ≤ 10μM
|
|
Z50266-1-O |
Enterococcus Faecium (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
750 |
0.24 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
5 |
0.33 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
60 |
0.29 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7000 |
0.21 |
Functional ≤ 10μM
|
|
Z80443-1-O |
S180 (Sarcoma Cells) (cluster #1 Of 1), Other |
Other |
1730 |
0.23 |
Functional ≤ 10μM
|
|
Z80830-1-O |
Ehrlich (Ehrlich Ascites Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1820 |
0.23 |
Functional ≤ 10μM
|
|
Z80860-1-O |
GC3/M Cell Line (cluster #1 Of 1), Other |
Other |
2130 |
0.23 |
Functional ≤ 10μM
|
|
Z80862-1-O |
GC3/MTK- Cell Line (cluster #1 Of 1), Other |
Other |
1400 |
0.23 |
Functional ≤ 10μM
|
|
Z80890-1-O |
H350.3 (MTX-resistant Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z81133-1-O |
L1210 (1565) (cluster #1 Of 1), Other |
Other |
5500 |
0.21 |
Functional ≤ 10μM
|
|
Z81221-1-O |
Lymphoma Cell Line (cluster #1 Of 1), Other |
Other |
560 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.82 |
10.16 |
-129.42 |
4 |
11 |
-2 |
184 |
475.461 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.36 |
8.12 |
-191.26 |
3 |
11 |
-3 |
187 |
474.453 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.82 |
8.22 |
-71.83 |
5 |
11 |
-1 |
182 |
476.469 |
10 |
↓
|
|