|
|
Analogs
-
3873971
-
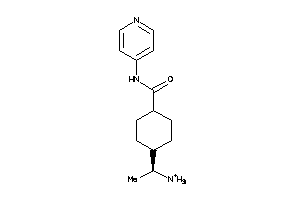
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCL2-1-E |
C-C Motif Chemokine 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2630 |
0.43 |
Binding ≤ 10μM
|
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
335 |
0.50 |
Binding ≤ 10μM
|
|
KS6A5-2-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8300 |
0.40 |
Binding ≤ 10μM
|
|
PKN2-1-E |
Protein Kinase N2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.56 |
Binding ≤ 10μM
|
|
PKN2-1-E |
Protein Kinase N2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.48 |
Binding ≤ 10μM
|
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
140 |
0.53 |
Binding ≤ 10μM
|
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
871 |
0.47 |
Binding ≤ 10μM
|
|
ROCK2-2-E |
Rho-associated Protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
245 |
0.51 |
Binding ≤ 10μM
|
|
ROCK2-2-E |
Rho-associated Protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.47 |
Binding ≤ 10μM
|
|
Z50597-6-O |
Rattus Norvegicus (cluster #6 Of 12), Other |
Other |
4750 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.72 |
3.52 |
-39.49 |
4 |
4 |
1 |
70 |
248.35 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.72 |
5.1 |
-91.09 |
7 |
8 |
2 |
122 |
438.507 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7762 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
1.86 |
-51.87 |
5 |
3 |
1 |
67 |
174.227 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.82 |
1.46 |
-6.23 |
4 |
3 |
0 |
65 |
173.219 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.82 |
2.32 |
-89.31 |
6 |
3 |
2 |
68 |
175.235 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3162 |
0.23 |
Binding ≤ 10μM
|
|
PDPK1-1-E |
3-phosphoinositide Dependent Protein Kinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
293 |
0.27 |
Binding ≤ 10μM
|
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7943 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
10.2 |
-41.93 |
6 |
9 |
1 |
126 |
463.61 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.11 |
10.07 |
-21.43 |
5 |
9 |
0 |
125 |
462.602 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 45 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCL2-1-E |
C-C Motif Chemokine 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6918 |
0.36 |
Binding ≤ 10μM
|
|
KAPCA-2-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
460 |
0.44 |
Binding ≤ 10μM
|
|
KAPCB-2-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.44 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
460 |
0.44 |
Binding ≤ 10μM
|
|
KPCE-4-E |
Protein Kinase C Epsilon (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM
|
|
KS6A5-1-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.37 |
Binding ≤ 10μM
|
|
MYLK-1-E |
Myosin Light Chain Kinase, Smooth Muscle (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.38 |
Binding ≤ 10μM
|
|
PKN1-1-E |
Protein Kinase N1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.40 |
Binding ≤ 10μM
|
|
PKN2-1-E |
Protein Kinase N2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.43 |
Binding ≤ 10μM
|
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
180 |
0.47 |
Binding ≤ 10μM
|
|
ROCK2-1-E |
Rho-associated Protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.40 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
7500 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
3.5 |
-58.49 |
2 |
5 |
1 |
67 |
292.384 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.88 |
2.33 |
-15.91 |
1 |
5 |
0 |
62 |
291.376 |
2 |
↓
|
|
|
|
Analogs
-
22708
-
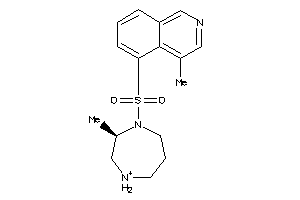
-
3940873
-
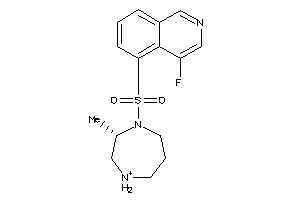
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.52 |
Binding ≤ 10μM
|
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.48 |
Binding ≤ 10μM
|
|
ROCK2-1-E |
Rho-associated Protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.50 |
Binding ≤ 10μM
|
|
Z80344-1-O |
NT2 (Teratocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
2500 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
4.88 |
-54.88 |
2 |
5 |
1 |
67 |
320.438 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.64 |
3.67 |
-14.03 |
1 |
5 |
0 |
62 |
319.43 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
790 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.06 |
4.46 |
-65.01 |
5 |
5 |
1 |
85 |
281.339 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.06 |
4.17 |
-14.03 |
4 |
5 |
0 |
84 |
280.331 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
14.9 |
-86.78 |
5 |
8 |
2 |
97 |
390.495 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.96 |
13.18 |
-8.44 |
3 |
8 |
0 |
91 |
388.479 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.97 |
13.64 |
-36.69 |
4 |
8 |
1 |
92 |
389.487 |
5 |
↓
|
|
|
|
Analogs
-
3940873
-

-
22706
-
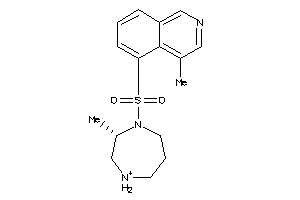
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
4.74 |
-55.11 |
2 |
5 |
1 |
67 |
320.438 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.64 |
3.44 |
-14.79 |
1 |
5 |
0 |
62 |
319.43 |
2 |
↓
|
|