|
|
Analogs
-
3873971
-
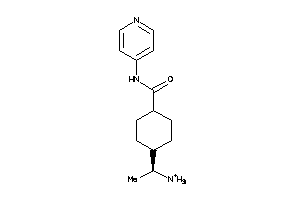
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCL2-1-E |
C-C Motif Chemokine 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2630 |
0.43 |
Binding ≤ 10μM
|
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
335 |
0.50 |
Binding ≤ 10μM
|
|
KS6A5-2-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8300 |
0.40 |
Binding ≤ 10μM
|
|
PKN2-1-E |
Protein Kinase N2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.56 |
Binding ≤ 10μM
|
|
PKN2-1-E |
Protein Kinase N2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.48 |
Binding ≤ 10μM
|
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
140 |
0.53 |
Binding ≤ 10μM
|
|
ROCK1-2-E |
Rho-associated Protein Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
871 |
0.47 |
Binding ≤ 10μM
|
|
ROCK2-2-E |
Rho-associated Protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
245 |
0.51 |
Binding ≤ 10μM
|
|
ROCK2-2-E |
Rho-associated Protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.47 |
Binding ≤ 10μM
|
|
Z50597-6-O |
Rattus Norvegicus (cluster #6 Of 12), Other |
Other |
4750 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.72 |
3.52 |
-39.49 |
4 |
4 |
1 |
70 |
248.35 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.72 |
5.1 |
-91.09 |
7 |
8 |
2 |
122 |
438.507 |
5 |
↓
|
|
|
|
Analogs
-
3965914
-
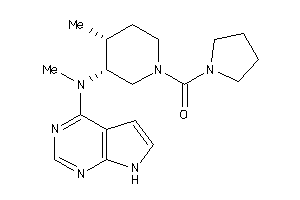
Draw
Identity
99%
90%
80%
70%
Vendors
And 36 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DCLK3-1-E |
Serine/threonine-protein Kinase DCLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
JAK1-1-E |
Tyrosine-protein Kinase JAK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM |
|
JAK2-1-E |
Tyrosine-protein Kinase JAK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
JAK3-2-E |
Tyrosine-protein Kinase JAK3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
48 |
0.45 |
Binding ≤ 10μM |
|
KCC1A-1-E |
CaM Kinase I Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.32 |
Binding ≤ 10μM
|
|
KS6A2-1-E |
Ribosomal Protein S6 Kinase Alpha 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.36 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3870 |
0.33 |
Binding ≤ 10μM
|
|
NUAK2-1-E |
NUAK Family SNF1-like Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.39 |
Binding ≤ 10μM
|
|
PKN1-1-E |
Protein Kinase N1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.41 |
Binding ≤ 10μM
|
|
ROCK2-2-E |
Rho-associated Protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.33 |
Binding ≤ 10μM
|
|
STK3-1-E |
Serine/threonine-protein Kinase MST2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4300 |
0.33 |
Binding ≤ 10μM
|
|
TNK1-1-E |
Non-receptor Tyrosine-protein Kinase TNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
640 |
0.38 |
Binding ≤ 10μM
|
|
TYK2-1-E |
Tyrosine-protein Kinase TYK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
620 |
0.38 |
Binding ≤ 10μM
|
|
Z80036-1-O |
BaF3 (IL-3-dependent Pro-B-cells) (cluster #1 Of 3), Other |
Other |
570 |
0.38 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
7840 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.45 |
9.01 |
-25.29 |
1 |
7 |
0 |
89 |
312.377 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.01 |
6.74 |
-49.17 |
1 |
7 |
1 |
86 |
313.385 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.45 |
9.48 |
-43.37 |
2 |
7 |
1 |
90 |
313.385 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ROCK2-2-E |
Rho-associated Protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
72 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
6.51 |
-17.24 |
2 |
6 |
0 |
76 |
321.336 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ROCK2-2-E |
Rho-associated Protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
72 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
6.5 |
-17.51 |
2 |
6 |
0 |
76 |
321.336 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7491 |
0.24 |
Binding ≤ 10μM
|
|
ROCK1-1-E |
Rho-associated Protein Kinase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.34 |
Binding ≤ 10μM
|
|
ROCK2-2-E |
Rho-associated Protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
8.27 |
-53.55 |
3 |
8 |
1 |
90 |
409.466 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.38 |
5.75 |
-14.73 |
2 |
8 |
0 |
89 |
408.458 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7491 |
0.24 |
Binding ≤ 10μM
|
|
ROCK1-1-E |
Rho-associated Protein Kinase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.34 |
Binding ≤ 10μM
|
|
ROCK2-2-E |
Rho-associated Protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
8.27 |
-51.81 |
3 |
8 |
1 |
90 |
409.466 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.38 |
5.75 |
-14.93 |
2 |
8 |
0 |
89 |
408.458 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KGP1-1-E |
CGMP-dependent Protein Kinase 1 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.23 |
Binding ≤ 10μM
|
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3000 |
0.23 |
Binding ≤ 10μM
|
|
PKN2-1-E |
Protein Kinase N2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
770 |
0.26 |
Binding ≤ 10μM
|
|
ROCK2-2-E |
Rho-associated Protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
3.37 |
-24.41 |
6 |
8 |
0 |
125 |
443.507 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.06 |
4.74 |
-66.45 |
7 |
8 |
1 |
130 |
444.515 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KGP1-1-E |
CGMP-dependent Protein Kinase 1 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5600 |
0.22 |
Binding ≤ 10μM
|
|
ROCK2-2-E |
Rho-associated Protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
33 |
0.32 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
6400 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
6.57 |
-19.54 |
3 |
7 |
0 |
89 |
445.519 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.57 |
7.94 |
-62.53 |
4 |
7 |
1 |
93 |
446.527 |
7 |
↓
|
|