|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.24 |
12.76 |
-58.2 |
3 |
6 |
1 |
96 |
475.609 |
13 |
↓
|
|
|
|
Analogs
-
2200
-
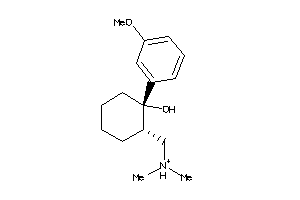
-
488880
-
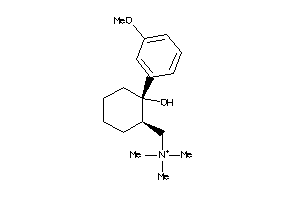
-
488881
-
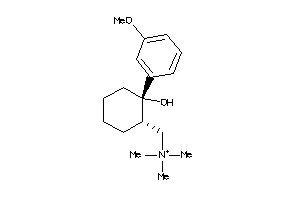
-
1849532
-
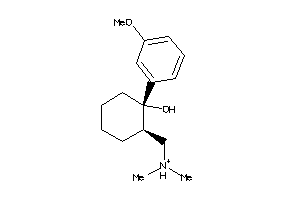
-
2015652
-
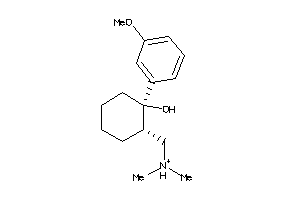
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.59 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
14 |
0.58 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5300 |
0.39 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7600 |
0.38 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
790 |
0.45 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.42 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.40 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.40 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1100 |
0.44 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1900 |
0.42 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3100 |
0.41 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4300 |
0.40 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu Opioid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
6.41 |
-36.54 |
2 |
3 |
1 |
34 |
264.389 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
4044297
-
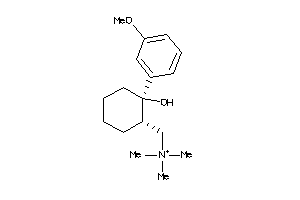
-
4044299
-
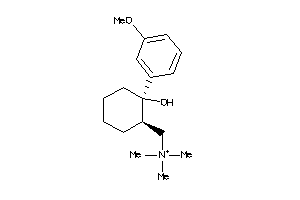
-
20781
-
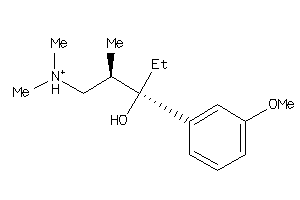
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
0.36 |
-34.37 |
2 |
3 |
1 |
33 |
264.389 |
4 |
↓
|
|
|
|
Analogs
-
4044297
-

-
4044299
-

-
20781
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
6.12 |
-31.63 |
2 |
3 |
1 |
34 |
264.389 |
4 |
↓
|
|
|
|
Analogs
-
3632418
-
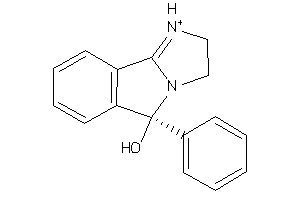
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
5.9 |
-28.14 |
2 |
3 |
1 |
37 |
251.309 |
1 |
↓
|
|
|
|
Analogs
-
3632416
-
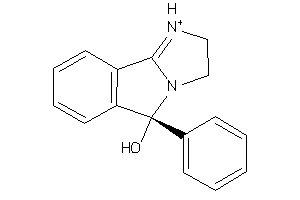
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
5.86 |
-28.29 |
2 |
3 |
1 |
37 |
251.309 |
1 |
↓
|
|
|
|
Analogs
-
2284
-
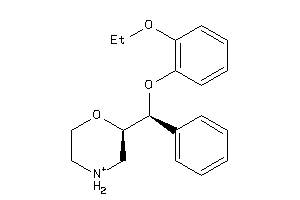
-
6923
-
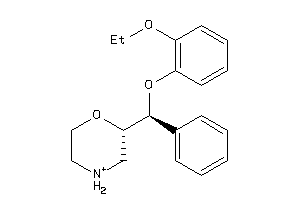
-
3996032
-
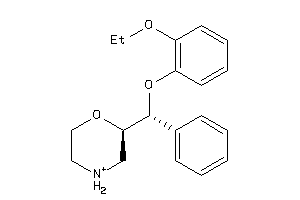
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.48 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.44 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
503 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
-1.24 |
-42.42 |
2 |
4 |
1 |
44 |
314.405 |
6 |
↓
|
|
|
|
Analogs
-
6923
-

-
3996032
-

-
2275
-
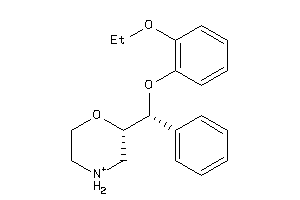
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
-1.11 |
-41.16 |
2 |
4 |
1 |
44 |
314.405 |
6 |
↓
|
|
|
|
Analogs
-
3996032
-

-
2275
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.48 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.44 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
503 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
7.92 |
-45.28 |
2 |
4 |
1 |
44 |
314.405 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.83 |
6.82 |
-8.25 |
1 |
4 |
0 |
40 |
313.397 |
6 |
↓
|
|
|
|
Analogs
-
2275
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.48 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.44 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
503 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
7.87 |
-46.18 |
2 |
4 |
1 |
44 |
314.405 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.83 |
6.8 |
-8.72 |
1 |
4 |
0 |
40 |
313.397 |
6 |
↓
|
|
|
|
Analogs
-
33895030
-
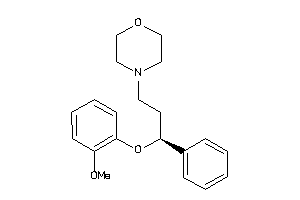
-
33895031
-
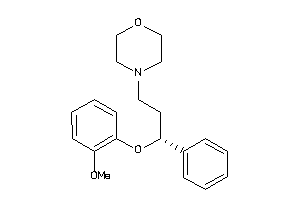
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
8.02 |
-42.17 |
2 |
3 |
1 |
35 |
272.368 |
7 |
↓
|
|
|
|
Analogs
-
33895030
-

-
33895031
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
8.02 |
-42.66 |
2 |
3 |
1 |
35 |
272.368 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9987 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
5.11 |
-42.27 |
3 |
1 |
1 |
28 |
196.339 |
2 |
↓
|
|
|
|
Analogs
-
28564671
-
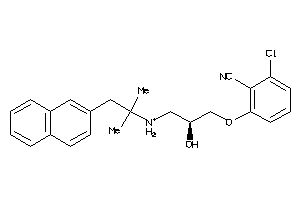
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.27 |
-0.48 |
-57.12 |
3 |
4 |
1 |
69 |
409.937 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.08 |
7.44 |
-94.67 |
7 |
7 |
2 |
113 |
432.572 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
787 |
0.43 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3608 |
0.38 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
5.75 |
-36.61 |
4 |
3 |
1 |
51 |
289.424 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1415 |
0.24 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Binding ≤ 10μM
|
|
CCR1-1-E |
C-C Chemokine Receptor Type 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Binding ≤ 10μM
|
|
CCR2-1-E |
C-C Chemokine Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Binding ≤ 10μM
|
|
CCR3-1-E |
C-C Chemokine Receptor Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.33 |
Binding ≤ 10μM
|
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Binding ≤ 10μM
|
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Binding ≤ 10μM
|
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
611 |
0.26 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
611 |
0.26 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
718 |
0.25 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2350 |
0.23 |
Binding ≤ 10μM
|
|
CCR3-1-E |
C-C Chemokine Receptor Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.33 |
Functional ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
240 |
0.27 |
ADME/T ≤ 10μM
|
|
DRD2-17-E |
Dopamine D2 Receptor (cluster #17 Of 24), Eukaryotic |
Eukaryotes |
321 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.40 |
14.84 |
-55.42 |
3 |
6 |
1 |
76 |
487.665 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
42 |
0.33 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.32 |
Binding ≤ 10μM
|
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
229 |
0.30 |
Binding ≤ 10μM
|
|
5HT6R-2-E |
Serotonin 6 (5-HT6) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.31 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.36 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
32 |
0.34 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
32 |
0.34 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
32 |
0.34 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1012 |
0.27 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
316 |
0.29 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
24 |
0.34 |
Binding ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
476 |
0.29 |
Binding ≤ 10μM
|
|
ADRB2-2-E |
Beta-2 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
61 |
0.33 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7396 |
0.23 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
63 |
0.33 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.36 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.31 |
Binding ≤ 10μM
|
|
HRH2-1-E |
Histamine H2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
302 |
0.29 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6980 |
0.23 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
407 |
0.29 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6144 |
0.24 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
480 |
0.29 |
Binding ≤ 10μM
|
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
DRD2-4-E |
Dopamine D2 Receptor (cluster #4 Of 24), Eukaryotic |
Eukaryotes |
22 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
-0.53 |
-48.34 |
1 |
8 |
1 |
73 |
424.525 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
4.25 |
-36.98 |
3 |
3 |
1 |
45 |
250.362 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
85 |
0.55 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
23 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
6.69 |
-37.46 |
2 |
3 |
1 |
39 |
328.617 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.36 |
5.17 |
-4.65 |
1 |
3 |
0 |
34 |
327.609 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.51 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6510 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.97 |
7.93 |
-35.86 |
2 |
2 |
1 |
26 |
266.723 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.97 |
6.38 |
-4.46 |
1 |
2 |
0 |
21 |
265.715 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
246 |
0.51 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
14 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
6.76 |
-41.81 |
2 |
3 |
1 |
39 |
263.748 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.00 |
5.26 |
-3.68 |
1 |
3 |
0 |
34 |
262.74 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.00 |
7.2 |
-106.9 |
3 |
3 |
2 |
40 |
264.756 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
8020 |
0.21 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
6280 |
0.21 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2830 |
0.23 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2470 |
0.23 |
Binding ≤ 10μM
|
|
SIRT1-1-E |
NAD-dependent Deacetylase Sirtuin 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
160 |
0.28 |
Binding ≤ 10μM
|
|
SIRT2-3-E |
NAD-dependent Deacetylase Sirtuin 2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
160 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
8.12 |
-58.41 |
3 |
8 |
1 |
92 |
470.582 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.59 |
3.63 |
-91.33 |
9 |
9 |
2 |
152 |
424.484 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
159 |
0.53 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
39 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
6.57 |
-37.99 |
2 |
3 |
1 |
39 |
284.166 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.23 |
5.05 |
-4.85 |
1 |
3 |
0 |
34 |
283.158 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
257 |
0.46 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
18 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
6.72 |
-37.32 |
2 |
4 |
1 |
48 |
273.356 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.62 |
5.21 |
-6.77 |
1 |
4 |
0 |
43 |
272.348 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.72 |
15.1 |
-38.71 |
1 |
3 |
1 |
17 |
415.601 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.72 |
12.86 |
-5.54 |
0 |
3 |
0 |
16 |
414.593 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7790 |
0.45 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9340 |
0.44 |
Binding ≤ 10μM
|
|
5HT6R-2-E |
Serotonin 6 (5-HT6) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1480 |
0.51 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5500 |
0.46 |
Binding ≤ 10μM
|
|
ADA2A-4-E |
Alpha-2a Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2540 |
0.49 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1130 |
0.52 |
Binding ≤ 10μM
|
|
ADA2C-4-E |
Alpha-2c Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
810 |
0.53 |
Binding ≤ 10μM
|
|
AOFA-4-E |
Monoamine Oxidase A (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
5 |
0.73 |
Binding ≤ 10μM
|
|
AOFB-6-E |
Monoamine Oxidase B (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
49 |
0.64 |
Binding ≤ 10μM
|
|
DYR1A-1-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.62 |
Binding ≤ 10μM
|
|
DYRK2-1-E |
Dual-specificity Tyrosine-phosphorylation Regulated Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
900 |
0.53 |
Binding ≤ 10μM
|
|
DYRK3-1-E |
Dual-specificity Tyrosine-phosphorylation Regulated Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.53 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.51 |
Binding ≤ 10μM
|
|
NISCH-3-E |
Nischarin (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
49 |
0.64 |
Binding ≤ 10μM
|
|
PIM3-1-E |
Serine/threonine-protein Kinase PIM3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.47 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3260 |
0.48 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
50 |
0.64 |
Functional ≤ 10μM
|
|
Z50607-3-O |
Human Immunodeficiency Virus 1 (cluster #3 Of 10), Other |
Other |
520 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
5.25 |
-32.65 |
2 |
3 |
1 |
39 |
213.26 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.63 |
4.83 |
-7.3 |
1 |
3 |
0 |
38 |
212.252 |
1 |
↓
|
|
|
|
Analogs
-
1999403
-
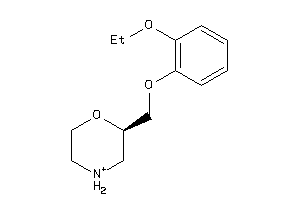
-
11587411
-
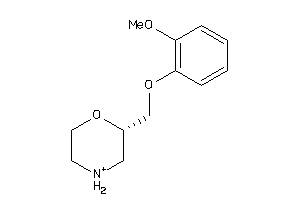
-
11587412
-
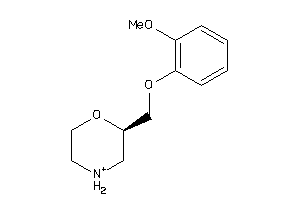
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
4.44 |
-46.92 |
2 |
4 |
1 |
44 |
238.307 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
13776229
-
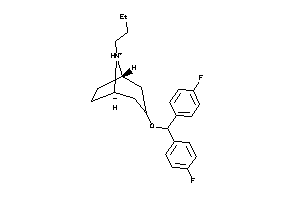
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.60 |
12.3 |
-38.72 |
1 |
2 |
1 |
14 |
344.425 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
38 |
0.55 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.80 |
7.3 |
-37.52 |
2 |
3 |
1 |
39 |
342.644 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.80 |
5.79 |
-4.37 |
1 |
3 |
0 |
34 |
341.636 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.80 |
7.69 |
-99.25 |
3 |
3 |
2 |
40 |
343.652 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
461 |
0.44 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2390 |
0.39 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
218 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
6.93 |
-37.18 |
2 |
4 |
1 |
48 |
273.356 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.57 |
5.42 |
-6.39 |
1 |
4 |
0 |
43 |
272.348 |
6 |
↓
|
|
|
|
Analogs
-
1530638
-
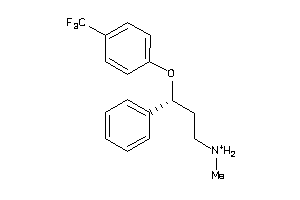
Draw
Identity
99%
90%
80%
70%
Vendors
And 51 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
ACES-10-E |
Acetylcholinesterase (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
130 |
0.44 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5820 |
0.33 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
176 |
0.43 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
563 |
0.40 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.51 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8300 |
0.32 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Functional ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
605 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3162 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.34 |
Functional ≤ 10μM
|
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
7000 |
0.33 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
33 |
0.48 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
8 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
9.16 |
-43.81 |
2 |
2 |
1 |
26 |
310.339 |
7 |
↓
|
|
|
|
Analogs
-
1530637
-
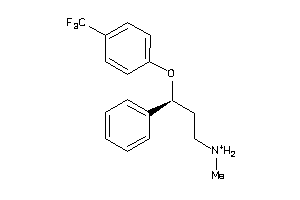
Draw
Identity
99%
90%
80%
70%
Vendors
And 50 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
ACES-10-E |
Acetylcholinesterase (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
130 |
0.44 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5820 |
0.33 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
176 |
0.43 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
563 |
0.40 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.51 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
31 |
0.48 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8300 |
0.32 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Functional ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
605 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3162 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.34 |
Functional ≤ 10μM
|
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
7000 |
0.33 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
21 |
0.49 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
8 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
9.16 |
-44.31 |
2 |
2 |
1 |
26 |
310.339 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
71312860
-
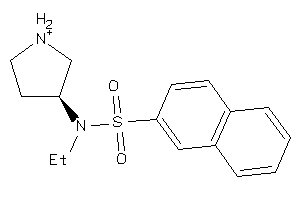
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.37 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
5.54 |
-49.49 |
2 |
4 |
1 |
54 |
305.423 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.71 |
4.1 |
-8.97 |
1 |
4 |
0 |
49 |
304.415 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.48 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.49 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
7.29 |
-40.82 |
2 |
2 |
1 |
26 |
228.315 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.48 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.49 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
7.26 |
-40.87 |
2 |
2 |
1 |
26 |
228.315 |
3 |
↓
|
|
|
|
Analogs
-
70519037
-
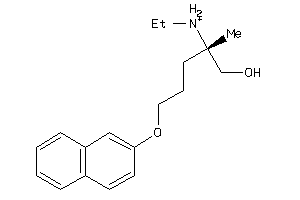
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
311 |
0.51 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3800 |
0.42 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.45 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
8 |
-42.18 |
2 |
2 |
1 |
26 |
242.342 |
3 |
↓
|
|
|
|
Analogs
-
35837228
-
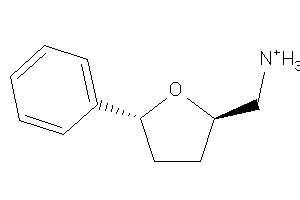
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2767 |
0.60 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
916 |
0.65 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.79 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.07 |
3.75 |
-41.25 |
3 |
2 |
1 |
37 |
178.255 |
2 |
↓
|
|
|
|
Analogs
-
35837226
-
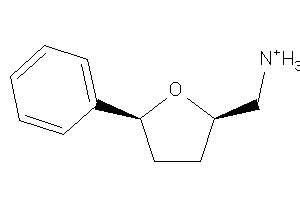
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2767 |
0.60 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
916 |
0.65 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.79 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.07 |
3.77 |
-40.97 |
3 |
2 |
1 |
37 |
178.255 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
40395186
-
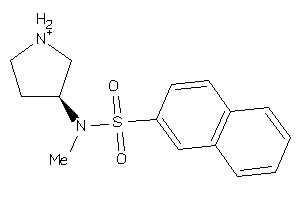
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
138 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.34 |
4.96 |
-49.8 |
2 |
4 |
1 |
54 |
291.396 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.34 |
3.49 |
-9.25 |
1 |
4 |
0 |
49 |
290.388 |
3 |
↓
|
|
|
|
Analogs
-
527387
-
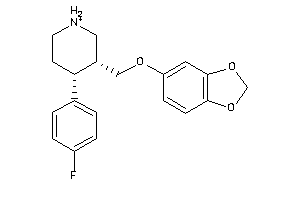
-
527385
-
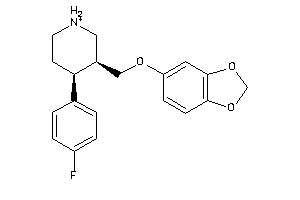
Draw
Identity
99%
90%
80%
70%
Vendors
And 61 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NORA-1-B |
Quinolone Resistance Protein NorA (cluster #1 Of 1), Bacterial |
Bacteria |
7000 |
0.30 |
Binding ≤ 10μM |
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6310 |
0.30 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6310 |
0.30 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6310 |
0.30 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
59 |
0.42 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.36 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.41 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.37 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.48 |
Functional ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4850 |
0.31 |
ADME/T ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
5320 |
0.31 |
Binding ≤ 10μM |
|
Z50185-3-O |
Staphylococcus Aureus (cluster #3 Of 4), Other |
Other |
10000 |
0.29 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.30 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
7620 |
0.30 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
7.73 |
-53.07 |
2 |
4 |
1 |
44 |
330.379 |
4 |
↓
|
|
|
|
Analogs
-
38450402
-
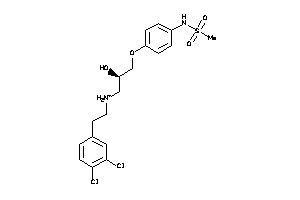
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
3.98 |
-66.19 |
4 |
6 |
1 |
92 |
434.365 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.10 |
2.75 |
-44.85 |
2 |
6 |
-1 |
90 |
432.349 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.10 |
4.15 |
-79.76 |
3 |
6 |
0 |
94 |
433.357 |
10 |
↓
|
|