|
|
Analogs
-
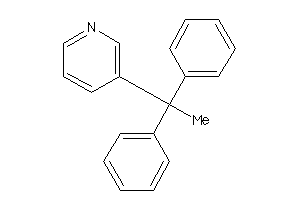
4051655
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7320 |
0.38 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
860 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
1.74 |
-4.44 |
0 |
1 |
0 |
12 |
245.325 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.86 |
1.86 |
-35.47 |
1 |
1 |
1 |
14 |
246.333 |
3 |
↓
|
|
|
|
Analogs
-
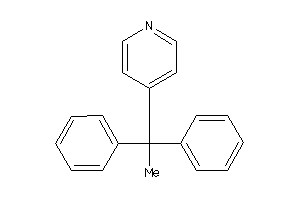
4051658
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3460 |
0.40 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
79 |
0.52 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1160 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
1.77 |
-4.61 |
0 |
1 |
0 |
12 |
245.325 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5761 |
0.41 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6043 |
0.41 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1517 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
-1 |
-41.87 |
3 |
1 |
1 |
28 |
240.37 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.72 |
15.1 |
-38.71 |
1 |
3 |
1 |
17 |
415.601 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.72 |
12.86 |
-5.54 |
0 |
3 |
0 |
16 |
414.593 |
10 |
↓
|
|
|
|
Analogs
-
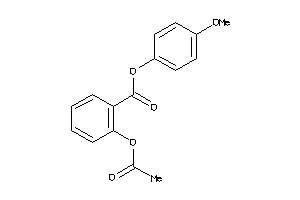
39280895
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
548 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
9.6 |
-13.21 |
0 |
4 |
0 |
53 |
256.257 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
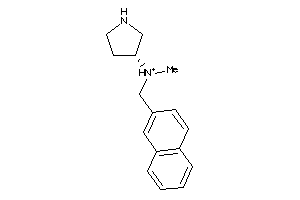
40892234
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5400 |
0.41 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7700 |
0.40 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
13 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
7.29 |
-37.3 |
2 |
2 |
1 |
16 |
241.358 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.83 |
6.45 |
-40.24 |
2 |
2 |
1 |
20 |
241.358 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.83 |
8.71 |
-114.56 |
3 |
2 |
2 |
21 |
242.366 |
3 |
↓
|
|
|
|
Analogs
-
34399350
-
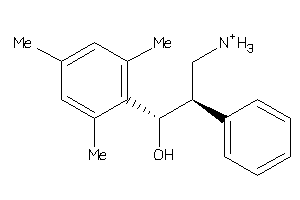
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8600 |
0.35 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
340 |
0.45 |
Binding ≤ 10μM
|
|
SC6A4-2-E |
Serotonin Transporter (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
643 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.20 |
6.56 |
-43.94 |
4 |
2 |
1 |
48 |
270.396 |
4 |
↓
|
|
|
|
Analogs
-
33852021
-
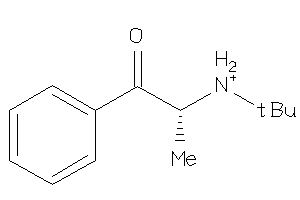
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.47 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5730 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.76 |
6.49 |
-34.87 |
2 |
2 |
1 |
34 |
206.309 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.76 |
5.33 |
-5.68 |
1 |
2 |
0 |
29 |
205.301 |
4 |
↓
|
|
|
|
Analogs
-
33852020
-
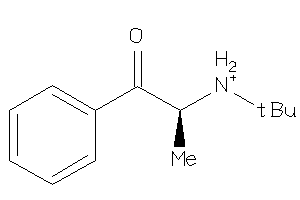
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.47 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5730 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.76 |
6.5 |
-34.93 |
2 |
2 |
1 |
34 |
206.309 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.76 |
5.43 |
-7.11 |
1 |
2 |
0 |
29 |
205.301 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
15.09 |
-44.26 |
1 |
3 |
1 |
17 |
449.565 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.07 |
12.77 |
-6.89 |
0 |
3 |
0 |
16 |
448.557 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.07 |
15.05 |
-42.25 |
1 |
3 |
1 |
17 |
449.565 |
9 |
↓
|
|
|
|
Analogs
-
3831139
-
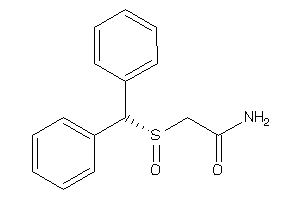
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3260 |
0.40 |
Binding ≤ 10μM |
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3800 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.22 |
4.11 |
-17.73 |
2 |
3 |
0 |
60 |
273.357 |
5 |
↓
|
|
|
|
Analogs
-
3875336
-
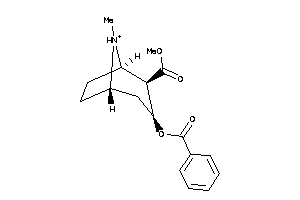
-
4214983
-
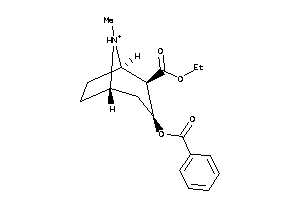
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
3.68 |
-33.02 |
1 |
5 |
1 |
57 |
318.393 |
6 |
↓
|
|
|
|
Analogs
-
4214983
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
3.43 |
-42.14 |
1 |
5 |
1 |
57 |
318.393 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2312 |
0.27 |
Binding ≤ 10μM
|
|
SC6A4-2-E |
Serotonin Transporter (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.66 |
15.78 |
-40.84 |
1 |
3 |
1 |
31 |
386.515 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.66 |
13.37 |
-6.75 |
0 |
3 |
0 |
30 |
385.507 |
6 |
↓
|
|
|
|
Analogs
-
33824876
-
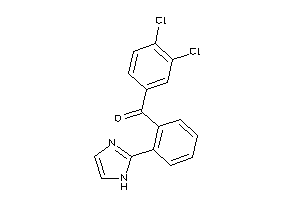
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2800 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.13 |
9.31 |
-12.19 |
1 |
3 |
0 |
46 |
282.73 |
3 |
↓
|
|
|
|
Analogs
-
6156
-
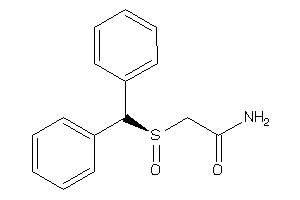
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3800 |
0.40 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7640 |
0.38 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.22 |
4.58 |
-19.45 |
2 |
3 |
0 |
60 |
273.357 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
124 |
0.44 |
Binding ≤ 10μM
|
|
SC6A4-2-E |
Serotonin Transporter (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9570 |
0.32 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1480 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
7.68 |
-38.71 |
2 |
3 |
1 |
29 |
297.422 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.80 |
10.02 |
-113.6 |
3 |
3 |
2 |
30 |
298.43 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.80 |
8.66 |
-33.13 |
2 |
3 |
1 |
26 |
297.422 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
651 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
8.75 |
-38.39 |
2 |
3 |
1 |
29 |
311.449 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.07 |
7.47 |
-4.08 |
1 |
3 |
0 |
24 |
310.441 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.07 |
9.51 |
-35.42 |
2 |
3 |
1 |
26 |
311.449 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
420 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
11.42 |
-35.57 |
1 |
2 |
1 |
14 |
282.407 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1570 |
0.41 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
0.11 |
-12.48 |
2 |
2 |
0 |
43 |
293.338 |
5 |
↓
|
|
|
|
Analogs
-
4214983
-

-
1319785
-
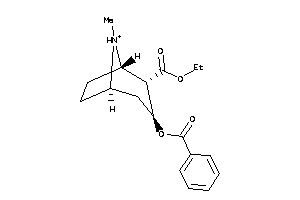
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9820 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
3.63 |
-32.13 |
1 |
5 |
1 |
57 |
304.366 |
5 |
↓
|
|
|
|
Analogs
-
39250338
-
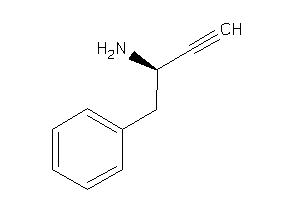
-
39250339
-
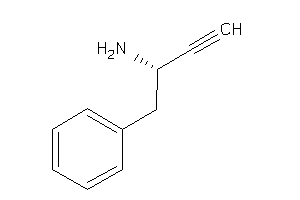
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7660 |
0.72 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7660 |
0.72 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7660 |
0.72 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7660 |
0.72 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Binding ≤ 10μM |
|
TAAR1-2-E |
Trace Amine-associated Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
920 |
0.85 |
Functional ≤ 10μM
|
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2850 |
0.78 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3500 |
0.76 |
ADME/T ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
490 |
0.88 |
Functional ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
800 |
0.85 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
4 |
-39.2 |
3 |
1 |
1 |
28 |
136.218 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
384061
-
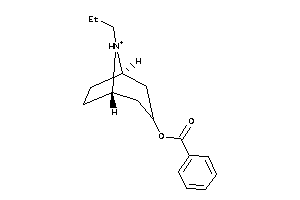
-
12410430
-
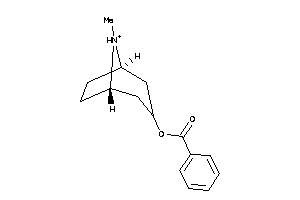
-
39291564
-
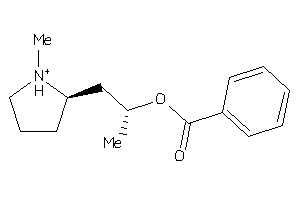
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
329 |
0.50 |
Binding ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
329 |
0.50 |
Binding ≤ 10μM
|
|
5HT3C-1-E |
Serotonin 3c (5-HT3c) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
329 |
0.50 |
Binding ≤ 10μM
|
|
5HT3D-1-E |
Serotonin 3d (5-HT3d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
329 |
0.50 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5180 |
0.41 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5180 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
2.32 |
-37.95 |
1 |
3 |
1 |
31 |
246.33 |
3 |
↓
|
|
|
|
Analogs
-
4214983
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-4-E |
HERG (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
7244 |
0.33 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9820 |
0.32 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
208 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
9.75 |
-41.49 |
1 |
5 |
1 |
57 |
304.366 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.87 |
7.41 |
-6.91 |
0 |
5 |
0 |
56 |
303.358 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
211 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
11.88 |
-31.58 |
1 |
5 |
1 |
57 |
332.42 |
6 |
↓
|
|