|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
91 |
0.66 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1425 |
0.55 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1425 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.08 |
-1.68 |
-13.7 |
7 |
7 |
0 |
134 |
229.265 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.21 |
-1.73 |
-13.45 |
7 |
7 |
0 |
137 |
229.265 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.08 |
-1.75 |
-14.11 |
7 |
7 |
0 |
134 |
229.265 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
203 |
0.62 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1215 |
0.55 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1215 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.65 |
1.07 |
-11.93 |
5 |
6 |
0 |
108 |
293.146 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.65 |
1.28 |
-41.01 |
4 |
6 |
-1 |
110 |
292.138 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.65 |
1.32 |
-38.67 |
4 |
6 |
-1 |
110 |
292.138 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
129 |
0.54 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1285 |
0.46 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1285 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.03 |
2.72 |
-13.47 |
5 |
6 |
0 |
108 |
264.31 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.03 |
2.92 |
-48.41 |
4 |
6 |
-1 |
110 |
263.302 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.03 |
2.96 |
-45.75 |
4 |
6 |
-1 |
110 |
263.302 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 44 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
95 |
0.70 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1350 |
0.59 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1350 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
-1.04 |
-14.13 |
6 |
6 |
0 |
125 |
214.25 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
4.68 |
-44.4 |
7 |
9 |
1 |
159 |
475.554 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
169 |
0.38 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1350 |
0.33 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1350 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
1.13 |
-13.32 |
2 |
8 |
0 |
110 |
361.427 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.82 |
0.54 |
-48.67 |
1 |
8 |
-1 |
107 |
360.419 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1230 |
0.59 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1230 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
1.71 |
-11.18 |
4 |
5 |
0 |
99 |
278.131 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
290 |
0.65 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1810 |
0.57 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1810 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.40 |
1.76 |
-13.17 |
4 |
5 |
0 |
99 |
213.262 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
170 |
0.56 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1235 |
0.49 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1235 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.22 |
1.16 |
-17.26 |
5 |
9 |
0 |
154 |
259.247 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.22 |
1.37 |
-26.06 |
5 |
9 |
0 |
158 |
259.247 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.35 |
1.27 |
-48.76 |
6 |
9 |
1 |
158 |
260.255 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
130 |
0.46 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1200 |
0.39 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.06 |
3.88 |
-13.14 |
5 |
7 |
0 |
111 |
307.379 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.06 |
4.21 |
-46.03 |
4 |
7 |
-1 |
113 |
306.371 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.06 |
4.18 |
-48.61 |
4 |
7 |
-1 |
113 |
306.371 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
225 |
0.66 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1170 |
0.59 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1170 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.63 |
1.6 |
-11.28 |
4 |
5 |
0 |
99 |
233.68 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
227 |
0.58 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1275 |
0.52 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1275 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.10 |
-0.32 |
-13.38 |
5 |
7 |
0 |
117 |
244.276 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.10 |
-0.05 |
-44.91 |
4 |
7 |
-1 |
119 |
243.268 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.10 |
-0.01 |
-42.45 |
4 |
7 |
-1 |
119 |
243.268 |
5 |
↓
|
|
|
|
Analogs
-
26481598
-
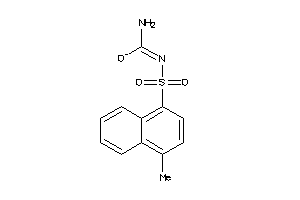
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
147 |
0.53 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1445 |
0.45 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1445 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.71 |
0.63 |
-11.08 |
2 |
5 |
0 |
82 |
264.306 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.71 |
0.05 |
-47.62 |
1 |
5 |
-1 |
79 |
263.298 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
240 |
0.62 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1320 |
0.55 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1320 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.01 |
0.38 |
-13.26 |
4 |
6 |
0 |
108 |
229.261 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
106 |
0.65 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1580 |
0.54 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1580 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.40 |
-3.77 |
-11.08 |
4 |
6 |
0 |
108 |
229.261 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.40 |
-4.36 |
-47.78 |
3 |
6 |
-1 |
105 |
228.253 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
185 |
0.79 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1320 |
0.69 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1320 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.10 |
-0.06 |
-47.12 |
3 |
5 |
-1 |
96 |
204.256 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
240 |
0.58 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1445 |
0.51 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1445 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.52 |
2.4 |
-59.56 |
4 |
7 |
-1 |
139 |
242.236 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
125 |
0.54 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1200 |
0.46 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.00 |
2.73 |
-14.1 |
5 |
6 |
0 |
108 |
264.31 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.00 |
2.96 |
-51.69 |
4 |
6 |
-1 |
110 |
263.302 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.00 |
2.66 |
-14.4 |
5 |
6 |
0 |
108 |
264.31 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
219 |
0.55 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1100 |
0.49 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.04 |
2.78 |
-11.78 |
5 |
6 |
0 |
108 |
256.331 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.04 |
3.01 |
-45.62 |
4 |
6 |
-1 |
110 |
255.323 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.04 |
3.05 |
-43.08 |
4 |
6 |
-1 |
110 |
255.323 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
146 |
0.53 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1350 |
0.46 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1350 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.48 |
1.56 |
-9.42 |
4 |
5 |
0 |
99 |
289.185 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
190 |
0.59 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1100 |
0.52 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.11 |
1.8 |
-15.42 |
4 |
8 |
0 |
144 |
244.232 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
325 |
0.53 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1550 |
0.48 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1550 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.05 |
-0.15 |
-55.79 |
2 |
7 |
-1 |
122 |
257.247 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.05 |
-0.74 |
-123.86 |
1 |
7 |
-2 |
119 |
256.239 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
180 |
0.59 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
975 |
0.53 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
975 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.09 |
1.8 |
-12.44 |
4 |
8 |
0 |
144 |
244.232 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
240 |
0.66 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1090 |
0.60 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1090 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.12 |
1.15 |
-11.43 |
4 |
5 |
0 |
99 |
217.225 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
195 |
0.55 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1070 |
0.49 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1070 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.83 |
-0.02 |
-18.83 |
5 |
7 |
0 |
128 |
256.287 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
139 |
0.38 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1355 |
0.33 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1355 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.14 |
3.14 |
-16.09 |
5 |
9 |
0 |
136 |
361.431 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.14 |
3.42 |
-49.75 |
4 |
9 |
-1 |
138 |
360.423 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.14 |
3.46 |
-47.14 |
4 |
9 |
-1 |
138 |
360.423 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
212 |
0.62 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1100 |
0.56 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.52 |
0.97 |
-11.99 |
5 |
6 |
0 |
108 |
248.695 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.52 |
1.18 |
-41.07 |
4 |
6 |
-1 |
110 |
247.687 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.52 |
1.21 |
-38.72 |
4 |
6 |
-1 |
110 |
247.687 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
123 |
0.51 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1350 |
0.43 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1350 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.37 |
0.9 |
-9.95 |
5 |
6 |
0 |
108 |
304.2 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.37 |
1.15 |
-34.19 |
4 |
6 |
-1 |
110 |
303.192 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
225 |
0.62 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1025 |
0.56 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1025 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.01 |
0.51 |
-12.33 |
5 |
6 |
0 |
108 |
232.24 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.12 |
0.75 |
-19.95 |
5 |
6 |
0 |
114 |
232.24 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.12 |
0.62 |
-41.2 |
6 |
6 |
1 |
112 |
233.248 |
3 |
↓
|
|
|
|
Analogs
-
12405017
-
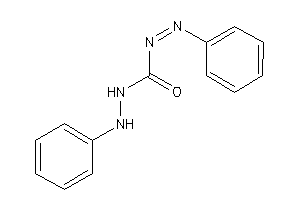
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
1700 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
-2.8 |
-11.33 |
2 |
5 |
0 |
65 |
240.266 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.72 |
3.62 |
-47.9 |
7 |
9 |
1 |
167 |
462.511 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
8800 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
2.29 |
-7.63 |
0 |
6 |
0 |
80 |
314.128 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ELNE-1-E |
Neutrophil Elastase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
24 |
0.63 |
Binding ≤ 10μM
|
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
680 |
0.51 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
170 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
2.32 |
-9.96 |
0 |
6 |
0 |
80 |
251.629 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ELNE-3-E |
Neutrophil Elastase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
28 |
0.62 |
Binding ≤ 10μM
|
|
KLKB1-1-E |
Plasma Kallikrein (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6100 |
0.43 |
Binding ≤ 10μM
|
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
4300 |
0.44 |
Binding ≤ 10μM
|
|
UROK-1-E |
Urokinase-type Plasminogen Activator (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3100 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.39 |
2.9 |
-6.86 |
0 |
6 |
0 |
80 |
231.211 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C1R-1-E |
Complement C1r (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1040 |
0.32 |
Binding ≤ 10μM |
|
C1S-1-E |
Complement C1s (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
140 |
0.37 |
Binding ≤ 10μM |
|
KLK1-1-E |
Kallikrein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM |
|
PLMN-1-E |
Plasminogen (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2900 |
0.30 |
Binding ≤ 10μM
|
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
290 |
0.35 |
Binding ≤ 10μM |
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
20 |
0.41 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
7.87 |
-80.13 |
9 |
7 |
2 |
142 |
349.394 |
6 |
↓
|
|