|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.73 |
7.42 |
-134.96 |
4 |
11 |
-2 |
184 |
493.451 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.27 |
6.33 |
-180.19 |
3 |
11 |
-3 |
187 |
492.443 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.73 |
6.3 |
-83.63 |
5 |
11 |
-1 |
182 |
494.459 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Binding ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
6000 |
0.37 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.29 |
0.57 |
-12.33 |
0 |
4 |
0 |
43 |
280.718 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.13 |
3.18 |
-145.4 |
4 |
13 |
-2 |
210 |
453.415 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.13 |
2.05 |
-85.51 |
5 |
13 |
-1 |
207 |
454.423 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
710 |
0.23 |
Binding ≤ 10μM |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
32 |
0.28 |
Binding ≤ 10μM |
|
E2QDN0-1-B |
Thymidylate Synthase (EC 2.1.1.45) (TS) (TSase) (cluster #1 Of 1), Bacterial |
Bacteria |
23 |
0.29 |
Binding ≤ 10μM |
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.03 |
12.11 |
-140.59 |
2 |
10 |
-2 |
158 |
498.495 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.03 |
10.13 |
-77.5 |
3 |
10 |
-1 |
156 |
499.503 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.26 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.26 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
5700 |
0.23 |
Binding ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
960 |
0.26 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
100 |
0.31 |
Functional ≤ 10μM |
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
6 |
0.36 |
Functional ≤ 10μM |
|
Z81133-1-O |
L1210 (1565) (cluster #1 Of 1), Other |
Other |
760 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.67 |
6.8 |
-126.25 |
2 |
10 |
-2 |
158 |
456.48 |
9 |
↓
|
|
|
|
Analogs
-
1851132
-
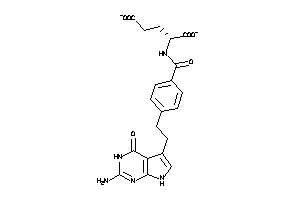
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.29 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
380 |
0.29 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
340 |
0.29 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
2300 |
0.25 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
1100 |
0.27 |
Binding ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
16 |
0.35 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
22 |
0.35 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
894 |
0.27 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
68 |
0.32 |
Functional ≤ 10μM |
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
894 |
0.27 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.53 |
8.7 |
-125.85 |
5 |
11 |
-2 |
197 |
425.401 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.53 |
6.72 |
-70.62 |
6 |
11 |
-1 |
194 |
426.409 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
22 |
0.31 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
90 |
0.28 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
95 |
0.28 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
430 |
0.25 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.28 |
Binding ≤ 10μM
|
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.21 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
360 |
0.26 |
Binding ≤ 10μM
|
|
Z50266-1-O |
Enterococcus Faecium (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
750 |
0.24 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
5 |
0.33 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7000 |
0.21 |
Functional ≤ 10μM
|
|
Z80443-1-O |
S180 (Sarcoma Cells) (cluster #1 Of 1), Other |
Other |
1730 |
0.23 |
Functional ≤ 10μM
|
|
Z80830-1-O |
Ehrlich (Ehrlich Ascites Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1820 |
0.23 |
Functional ≤ 10μM
|
|
Z80860-1-O |
GC3/M Cell Line (cluster #1 Of 1), Other |
Other |
2130 |
0.23 |
Functional ≤ 10μM
|
|
Z80862-1-O |
GC3/MTK- Cell Line (cluster #1 Of 1), Other |
Other |
1400 |
0.23 |
Functional ≤ 10μM
|
|
Z80890-1-O |
H350.3 (MTX-resistant Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z81221-1-O |
Lymphoma Cell Line (cluster #1 Of 1), Other |
Other |
560 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.82 |
8.37 |
-132.31 |
4 |
11 |
-2 |
184 |
475.461 |
10 |
↓
|
|
|
|
|
|
|
Analogs
-
4228260
-
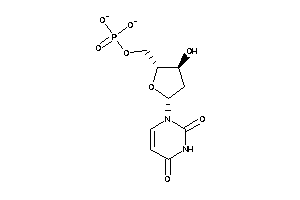
-
1532629
-
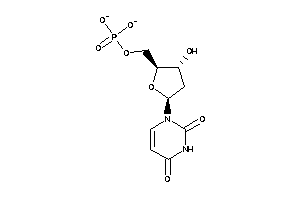
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
2300 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.80 |
-6.32 |
-137.45 |
2 |
10 |
-2 |
156 |
306.167 |
4 |
↓
|
|
|
|
Analogs
-
4228260
-

-
1532629
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
2300 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.80 |
-5.85 |
-149.9 |
2 |
10 |
-2 |
156 |
306.167 |
4 |
↓
|
|
|
|
Analogs
-
4228260
-

-
1532629
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
2300 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.80 |
-6.14 |
-156.3 |
2 |
10 |
-2 |
156 |
306.167 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
-3.25 |
-12.39 |
3 |
5 |
0 |
84 |
284.344 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C3SWJ7-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
19 |
0.31 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
22 |
0.31 |
Binding ≤ 10μM
|
|
E2QDN0-1-B |
Thymidylate Synthase (EC 2.1.1.45) (TS) (TSase) (cluster #1 Of 1), Bacterial |
Bacteria |
48 |
0.29 |
Binding ≤ 10μM |
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
90 |
0.28 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
95 |
0.28 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.29 |
Binding ≤ 10μM |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
430 |
0.25 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.28 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.28 |
Binding ≤ 10μM
|
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.21 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
360 |
0.26 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
360 |
0.26 |
Binding ≤ 10μM
|
|
Z100337-1-O |
W1-L2 (cluster #1 Of 1), Other |
Other |
440 |
0.25 |
Functional ≤ 10μM
|
|
Z50266-1-O |
Enterococcus Faecium (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
750 |
0.24 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
5 |
0.33 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
60 |
0.29 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7000 |
0.21 |
Functional ≤ 10μM
|
|
Z80443-1-O |
S180 (Sarcoma Cells) (cluster #1 Of 1), Other |
Other |
1730 |
0.23 |
Functional ≤ 10μM
|
|
Z80830-1-O |
Ehrlich (Ehrlich Ascites Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1820 |
0.23 |
Functional ≤ 10μM
|
|
Z80860-1-O |
GC3/M Cell Line (cluster #1 Of 1), Other |
Other |
2130 |
0.23 |
Functional ≤ 10μM
|
|
Z80862-1-O |
GC3/MTK- Cell Line (cluster #1 Of 1), Other |
Other |
1400 |
0.23 |
Functional ≤ 10μM
|
|
Z80890-1-O |
H350.3 (MTX-resistant Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z81133-1-O |
L1210 (1565) (cluster #1 Of 1), Other |
Other |
5500 |
0.21 |
Functional ≤ 10μM
|
|
Z81221-1-O |
Lymphoma Cell Line (cluster #1 Of 1), Other |
Other |
560 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.82 |
10.16 |
-129.42 |
4 |
11 |
-2 |
184 |
475.461 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.36 |
8.12 |
-191.26 |
3 |
11 |
-3 |
187 |
474.453 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.82 |
8.22 |
-71.83 |
5 |
11 |
-1 |
182 |
476.469 |
10 |
↓
|
|
|
|
|
|
|
Analogs
-
1529898
-
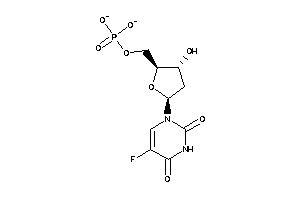
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
-5.84 |
-140.72 |
2 |
10 |
-2 |
156 |
324.157 |
4 |
↓
|
|
|
|
Analogs
-
1529898
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
-4.64 |
-163.61 |
2 |
10 |
-2 |
156 |
324.157 |
4 |
↓
|
|
|
|
Analogs
-
1529898
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
-5.54 |
-139.88 |
2 |
10 |
-2 |
156 |
324.157 |
4 |
↓
|
|
|
|
Analogs
-
6920406
-
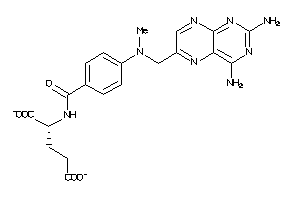
Draw
Identity
99%
90%
80%
70%
Vendors
And 72 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B0BL08-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.34 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
7 |
0.35 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
9 |
0.34 |
Binding ≤ 10μM
|
|
DYR1-1-B |
Dihydrofolate Reductase Type 1 (cluster #1 Of 1), Bacterial |
Bacteria |
7 |
0.35 |
Binding ≤ 10μM
|
|
DYR1-1-B |
Dihydrofolate Reductase Type 1 (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.34 |
Binding ≤ 10μM
|
|
Q81R22-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
15 |
0.33 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
1800 |
0.24 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
26 |
0.32 |
Functional ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.34 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
33 |
0.32 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.35 |
Binding ≤ 10μM
|
|
FOLC-1-E |
Folylpoly-gamma-glutamate Synthetase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.23 |
Binding ≤ 10μM
|
|
PTR1-1-E |
Pteridine Reductase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
39 |
0.31 |
Binding ≤ 10μM
|
|
S19A1-1-E |
Folate Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.23 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3600 |
0.23 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
880 |
0.26 |
Binding ≤ 10μM
|
|
Z103204-2-O |
A427 (cluster #2 Of 4), Other |
Other |
5520 |
0.22 |
Functional ≤ 10μM
|
|
Z50266-1-O |
Enterococcus Faecium (cluster #1 Of 1), Other |
Other |
90 |
0.30 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
85 |
0.30 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
6600 |
0.22 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
4200 |
0.23 |
Functional ≤ 10μM
|
|
Z80001-1-O |
143B (cluster #1 Of 2), Other |
Other |
9 |
0.34 |
Functional ≤ 10μM
|
|
Z80008-1-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
16 |
0.33 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
40 |
0.31 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
1100 |
0.25 |
Functional ≤ 10μM
|
|
Z80063-1-O |
CCRF S-180 (cluster #1 Of 1), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
620 |
0.26 |
Functional ≤ 10μM
|
|
Z80099-1-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
870 |
0.26 |
Functional ≤ 10μM
|
|
Z80112-1-O |
DAN-G (cluster #1 Of 2), Other |
Other |
77 |
0.30 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
45 |
0.31 |
Functional ≤ 10μM
|
|
Z80132-1-O |
EL4 (Thymoma Cells) (cluster #1 Of 2), Other |
Other |
1570 |
0.25 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7 |
0.35 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z80166-12-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #12 Of 12), Other |
Other |
4400 |
0.23 |
Functional ≤ 10μM |
|
Z80171-1-O |
HuTu80 (cluster #1 Of 1), Other |
Other |
9 |
0.34 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
26 |
0.32 |
Functional ≤ 10μM
|
|
Z80192-1-O |
KM12 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
42 |
0.31 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z80214-1-O |
M14 (Melanoma Cells) (cluster #1 Of 3), Other |
Other |
32 |
0.32 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
900 |
0.26 |
Functional ≤ 10μM
|
|
Z80227-1-O |
MCF7-ADR (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
78 |
0.30 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
28 |
0.32 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
400 |
0.27 |
Functional ≤ 10μM
|
|
Z80356-1-O |
OVCAR-5 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
980 |
0.25 |
Functional ≤ 10μM
|
|
Z80357-1-O |
OVCAR-8 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
31 |
0.32 |
Functional ≤ 10μM
|
|
Z80362-8-O |
P388 (Lymphoma Cells) (cluster #8 Of 8), Other |
Other |
66 |
0.30 |
Functional ≤ 10μM |
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
27 |
0.32 |
Functional ≤ 10μM
|
|
Z80407-1-O |
R1 (cluster #1 Of 1), Other |
Other |
720 |
0.26 |
Functional ≤ 10μM
|
|
Z80408-1-O |
R1-CCRF-CEM (Lymphoblastic Leukemia) (cluster #1 Of 1), Other |
Other |
985 |
0.25 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
216 |
0.28 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
2600 |
0.24 |
Functional ≤ 10μM
|
|
Z80410-1-O |
R2-CCRF-CEM (Lymphoblastic Leukemia) (cluster #1 Of 1), Other |
Other |
2630 |
0.24 |
Functional ≤ 10μM
|
|
Z80411-1-O |
R30dm-CCRF-CEM (Lymphoblastic Leukemia) (cluster #1 Of 1), Other |
Other |
18 |
0.33 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
40 |
0.31 |
Functional ≤ 10μM
|
|
Z80433-1-O |
RPMI-8226 (Multiple Myeloma Cells) (cluster #1 Of 3), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z80443-1-O |
S180 (Sarcoma Cells) (cluster #1 Of 1), Other |
Other |
5 |
0.35 |
Functional ≤ 10μM
|
|
Z80455-1-O |
SCC-15 (cluster #1 Of 1), Other |
Other |
580 |
0.26 |
Functional ≤ 10μM
|
|
Z80457-1-O |
SCC-25 (cluster #1 Of 1), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z80459-1-O |
SCC-7 (cluster #1 Of 1), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM
|
|
Z80468-1-O |
SF-295 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
36 |
0.32 |
Functional ≤ 10μM
|
|
Z80488-2-O |
SK-MEL-5 (Melanoma Cells) (cluster #2 Of 3), Other |
Other |
87 |
0.30 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
15 |
0.33 |
Functional ≤ 10μM
|
|
Z80560-1-O |
U-251 (Glioma Cells) (cluster #1 Of 3), Other |
Other |
63 |
0.31 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
12 |
0.34 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
22 |
0.32 |
Functional ≤ 10μM
|
|
Z80568-1-O |
UACC-257 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
790 |
0.26 |
Functional ≤ 10μM
|
|
Z80570-3-O |
UACC-62 (Melanoma Cells) (cluster #3 Of 3), Other |
Other |
28 |
0.32 |
Functional ≤ 10μM
|
|
Z80575-1-O |
UO-31 (Renal Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
191 |
0.29 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
9 |
0.34 |
Functional ≤ 10μM
|
|
Z80640-1-O |
786-0 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z80657-1-O |
A253 Cell Line (cluster #1 Of 1), Other |
Other |
3000 |
0.23 |
Functional ≤ 10μM
|
|
Z80682-11-O |
A549 (Lung Carcinoma Cells) (cluster #11 Of 11), Other |
Other |
5 |
0.35 |
Functional ≤ 10μM |
|
Z80682-11-O |
A549 (Lung Carcinoma Cells) (cluster #11 Of 11), Other |
Other |
37 |
0.32 |
Functional ≤ 10μM |
|
Z80711-1-O |
NCI/ADR-RES (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
78 |
0.30 |
Functional ≤ 10μM
|
|
Z80752-1-O |
Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
15 |
0.33 |
Functional ≤ 10μM
|
|
Z80789-1-O |
Colon 38 (cluster #1 Of 1), Other |
Other |
66 |
0.30 |
Functional ≤ 10μM
|
|
Z80815-1-O |
D54 Cell Line (cluster #1 Of 1), Other |
Other |
16 |
0.33 |
Functional ≤ 10μM
|
|
Z80830-1-O |
Ehrlich (Ehrlich Ascites Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
13 |
0.33 |
Functional ≤ 10μM
|
|
Z80847-1-O |
FaDu (Pharyngeal Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
31 |
0.32 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
9740 |
0.21 |
Functional ≤ 10μM
|
|
Z80890-1-O |
H350.3 (MTX-resistant Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
906 |
0.26 |
Functional ≤ 10μM
|
|
Z80893-1-O |
H35 (Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z80920-1-O |
HCC 2998 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
110 |
0.30 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
30 |
0.32 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
25 |
0.32 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z81054-1-O |
SF-268 (Glioblastoma Cells) (cluster #1 Of 3), Other |
Other |
52 |
0.31 |
Functional ≤ 10μM
|
|
Z81065-1-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
65 |
0.30 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM |
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
26 |
0.32 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
100 |
0.30 |
Functional ≤ 10μM
|
|
Z81281-1-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 5), Other |
Other |
43 |
0.31 |
Functional ≤ 10μM
|
|
Z81283-1-O |
NCI-H522 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
450 |
0.27 |
Functional ≤ 10μM
|
|
Z81322-1-O |
SF-539 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
35 |
0.32 |
Functional ≤ 10μM
|
|
Z81325-1-O |
SR (Leukemia Cells) (cluster #1 Of 2), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
30 |
0.32 |
Functional ≤ 10μM
|
|
Z80253-1-O |
MEF (Embryonic Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
120 |
0.29 |
ADME/T ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
216 |
0.28 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.97 |
6.06 |
-125.84 |
5 |
13 |
-2 |
216 |
452.431 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.97 |
6.52 |
-124.82 |
6 |
13 |
-1 |
217 |
453.439 |
9 |
↓
|
|
|
|
Analogs
-
1529323
-
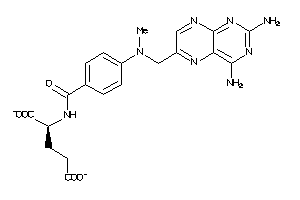
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B0BL08-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.34 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
9 |
0.34 |
Binding ≤ 10μM
|
|
DYR1-1-B |
Dihydrofolate Reductase Type 1 (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.34 |
Binding ≤ 10μM
|
|
Q81R22-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
15 |
0.33 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
1800 |
0.24 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
26 |
0.32 |
Functional ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
33 |
0.32 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.35 |
Binding ≤ 10μM
|
|
FOLC-1-E |
Folylpoly-gamma-glutamate Synthetase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.23 |
Binding ≤ 10μM
|
|
PTR1-1-E |
Pteridine Reductase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
39 |
0.31 |
Binding ≤ 10μM
|
|
S19A1-1-E |
Folate Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.23 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3600 |
0.23 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
880 |
0.26 |
Binding ≤ 10μM
|
|
Z103204-2-O |
A427 (cluster #2 Of 4), Other |
Other |
5520 |
0.22 |
Functional ≤ 10μM
|
|
Z50266-1-O |
Enterococcus Faecium (cluster #1 Of 1), Other |
Other |
90 |
0.30 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
85 |
0.30 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
6600 |
0.22 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
4200 |
0.23 |
Functional ≤ 10μM
|
|
Z80001-1-O |
143B (cluster #1 Of 2), Other |
Other |
9 |
0.34 |
Functional ≤ 10μM
|
|
Z80008-1-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
16 |
0.33 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
40 |
0.31 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
1100 |
0.25 |
Functional ≤ 10μM
|
|
Z80063-1-O |
CCRF S-180 (cluster #1 Of 1), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z80099-1-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
870 |
0.26 |
Functional ≤ 10μM
|
|
Z80112-1-O |
DAN-G (cluster #1 Of 2), Other |
Other |
77 |
0.30 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
45 |
0.31 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z80166-12-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #12 Of 12), Other |
Other |
4400 |
0.23 |
Functional ≤ 10μM |
|
Z80171-1-O |
HuTu80 (cluster #1 Of 1), Other |
Other |
9 |
0.34 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
26 |
0.32 |
Functional ≤ 10μM
|
|
Z80192-1-O |
KM12 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
42 |
0.31 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z80214-1-O |
M14 (Melanoma Cells) (cluster #1 Of 3), Other |
Other |
32 |
0.32 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
900 |
0.26 |
Functional ≤ 10μM
|
|
Z80227-1-O |
MCF7-ADR (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
78 |
0.30 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
28 |
0.32 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
400 |
0.27 |
Functional ≤ 10μM
|
|
Z80356-1-O |
OVCAR-5 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
980 |
0.25 |
Functional ≤ 10μM
|
|
Z80357-1-O |
OVCAR-8 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
31 |
0.32 |
Functional ≤ 10μM
|
|
Z80362-8-O |
P388 (Lymphoma Cells) (cluster #8 Of 8), Other |
Other |
66 |
0.30 |
Functional ≤ 10μM |
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
27 |
0.32 |
Functional ≤ 10μM
|
|
Z80407-1-O |
R1 (cluster #1 Of 1), Other |
Other |
720 |
0.26 |
Functional ≤ 10μM
|
|
Z80408-1-O |
R1-CCRF-CEM (Lymphoblastic Leukemia) (cluster #1 Of 1), Other |
Other |
985 |
0.25 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
2600 |
0.24 |
Functional ≤ 10μM
|
|
Z80410-1-O |
R2-CCRF-CEM (Lymphoblastic Leukemia) (cluster #1 Of 1), Other |
Other |
2630 |
0.24 |
Functional ≤ 10μM
|
|
Z80411-1-O |
R30dm-CCRF-CEM (Lymphoblastic Leukemia) (cluster #1 Of 1), Other |
Other |
18 |
0.33 |
Functional ≤ 10μM
|
|
Z80433-1-O |
RPMI-8226 (Multiple Myeloma Cells) (cluster #1 Of 3), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z80443-1-O |
S180 (Sarcoma Cells) (cluster #1 Of 1), Other |
Other |
5 |
0.35 |
Functional ≤ 10μM
|
|
Z80455-1-O |
SCC-15 (cluster #1 Of 1), Other |
Other |
580 |
0.26 |
Functional ≤ 10μM
|
|
Z80457-1-O |
SCC-25 (cluster #1 Of 1), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z80459-1-O |
SCC-7 (cluster #1 Of 1), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM
|
|
Z80468-1-O |
SF-295 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
36 |
0.32 |
Functional ≤ 10μM
|
|
Z80488-2-O |
SK-MEL-5 (Melanoma Cells) (cluster #2 Of 3), Other |
Other |
87 |
0.30 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
15 |
0.33 |
Functional ≤ 10μM
|
|
Z80560-1-O |
U-251 (Glioma Cells) (cluster #1 Of 3), Other |
Other |
63 |
0.31 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
12 |
0.34 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
22 |
0.32 |
Functional ≤ 10μM
|
|
Z80568-1-O |
UACC-257 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
790 |
0.26 |
Functional ≤ 10μM
|
|
Z80570-3-O |
UACC-62 (Melanoma Cells) (cluster #3 Of 3), Other |
Other |
28 |
0.32 |
Functional ≤ 10μM
|
|
Z80575-1-O |
UO-31 (Renal Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
191 |
0.29 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
9 |
0.34 |
Functional ≤ 10μM
|
|
Z80640-1-O |
786-0 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z80657-1-O |
A253 Cell Line (cluster #1 Of 1), Other |
Other |
3000 |
0.23 |
Functional ≤ 10μM
|
|
Z80682-11-O |
A549 (Lung Carcinoma Cells) (cluster #11 Of 11), Other |
Other |
5 |
0.35 |
Functional ≤ 10μM |
|
Z80711-1-O |
NCI/ADR-RES (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
78 |
0.30 |
Functional ≤ 10μM
|
|
Z80752-1-O |
Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
15 |
0.33 |
Functional ≤ 10μM
|
|
Z80789-1-O |
Colon 38 (cluster #1 Of 1), Other |
Other |
66 |
0.30 |
Functional ≤ 10μM
|
|
Z80815-1-O |
D54 Cell Line (cluster #1 Of 1), Other |
Other |
16 |
0.33 |
Functional ≤ 10μM
|
|
Z80830-1-O |
Ehrlich (Ehrlich Ascites Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
13 |
0.33 |
Functional ≤ 10μM
|
|
Z80847-1-O |
FaDu (Pharyngeal Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
31 |
0.32 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
9740 |
0.21 |
Functional ≤ 10μM
|
|
Z80890-1-O |
H350.3 (MTX-resistant Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
906 |
0.26 |
Functional ≤ 10μM
|
|
Z80893-1-O |
H35 (Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z80920-1-O |
HCC 2998 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
110 |
0.30 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
30 |
0.32 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
25 |
0.32 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z81054-1-O |
SF-268 (Glioblastoma Cells) (cluster #1 Of 3), Other |
Other |
52 |
0.31 |
Functional ≤ 10μM
|
|
Z81065-1-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
65 |
0.30 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM |
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
26 |
0.32 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
100 |
0.30 |
Functional ≤ 10μM
|
|
Z81281-1-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 5), Other |
Other |
43 |
0.31 |
Functional ≤ 10μM
|
|
Z81283-1-O |
NCI-H522 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
450 |
0.27 |
Functional ≤ 10μM
|
|
Z81322-1-O |
SF-539 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
35 |
0.32 |
Functional ≤ 10μM
|
|
Z81325-1-O |
SR (Leukemia Cells) (cluster #1 Of 2), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
30 |
0.32 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
216 |
0.28 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.97 |
6.03 |
-126.53 |
5 |
13 |
-2 |
216 |
452.431 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.97 |
6.57 |
-124.3 |
6 |
13 |
-1 |
217 |
453.439 |
9 |
↓
|
|
|
|
Analogs
-
8613263
-
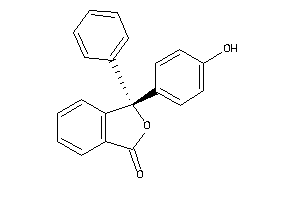
Draw
Identity
99%
90%
80%
70%
Vendors
And 40 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.35 |
Binding ≤ 10μM |
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
1400 |
0.34 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
5.89 |
-14.88 |
2 |
4 |
0 |
67 |
318.328 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.36 |
7.79 |
-128.62 |
2 |
12 |
-2 |
171 |
530.52 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.10 |
6.78 |
-185.31 |
1 |
12 |
-3 |
174 |
529.512 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.36 |
7.92 |
-80.53 |
3 |
12 |
-1 |
173 |
531.528 |
10 |
↓
|
|
|
|
Analogs
-
6523446
-
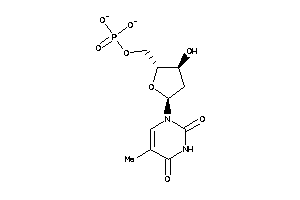
-
1532628
-
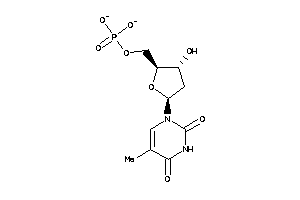
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.34 |
Binding ≤ 10μM
|
|
KTHY-1-B |
Thymidylate Kinase (cluster #1 Of 1), Bacterial |
Bacteria |
4500 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.10 |
-5.78 |
-152.67 |
2 |
10 |
-2 |
156 |
320.194 |
4 |
↓
|
|
|
|
Analogs
-
1532628
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.10 |
-1.51 |
-160.3 |
2 |
10 |
-2 |
157 |
320.194 |
4 |
↓
|
|
|
|
Analogs
-
6090961
-
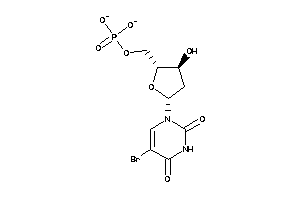
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.74 |
-6.13 |
-163.05 |
2 |
10 |
-2 |
156 |
385.063 |
4 |
↓
|
|
|
|
Analogs
-
6090961
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.74 |
-7.03 |
-138.18 |
2 |
10 |
-2 |
156 |
385.063 |
4 |
↓
|
|
|
|
Analogs
-
6090961
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.74 |
-7.2 |
-133.64 |
2 |
10 |
-2 |
156 |
385.063 |
4 |
↓
|
|
|
|
Analogs
-
6090961
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.74 |
-7.33 |
-139.65 |
2 |
10 |
-2 |
156 |
385.063 |
4 |
↓
|
|
|
|
Analogs
-
21289577
-
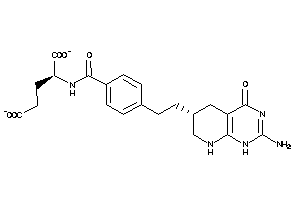
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
60 |
0.32 |
Binding ≤ 10μM |
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.30 |
Binding ≤ 10μM |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.33 |
Binding ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
59 |
0.32 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
15 |
0.34 |
Functional ≤ 10μM |
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
16 |
0.34 |
Functional ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
18 |
0.34 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
1500 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.12 |
5.35 |
-138.84 |
5 |
11 |
-2 |
193 |
441.444 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.12 |
4.23 |
-86.52 |
6 |
11 |
-1 |
190 |
442.452 |
9 |
↓
|
|
|
|
Analogs
-
12504292
-
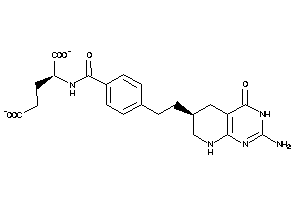
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.30 |
Binding ≤ 10μM |
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.27 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.33 |
Binding ≤ 10μM
|
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.30 |
Functional ≤ 10μM |
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
59 |
0.32 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
8 |
0.35 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
16 |
0.34 |
Functional ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
18 |
0.34 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
1500 |
0.25 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
1500 |
0.25 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
38 |
0.32 |
Functional ≤ 10μM |
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
31 |
0.33 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
31 |
0.33 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.12 |
8.18 |
-127.86 |
5 |
11 |
-2 |
193 |
441.444 |
9 |
↓
|
|
Ref
Reference (pH 7)
|
-1.12 |
7.51 |
-139.85 |
5 |
11 |
-2 |
193 |
441.444 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.12 |
5.53 |
-83.64 |
6 |
11 |
-1 |
190 |
442.452 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
670 |
0.27 |
Binding ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.67 |
6.8 |
-126.16 |
2 |
10 |
-2 |
158 |
456.48 |
9 |
↓
|
|
|
|
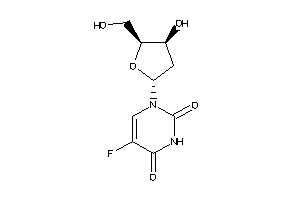
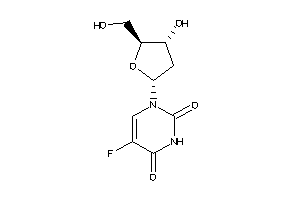
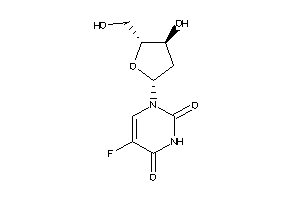
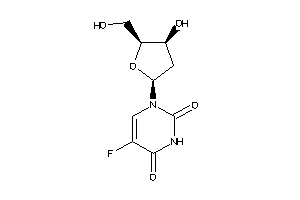
Analogs
-
388678
-
-
489135
-
-
3813010
-
-
3830846
-
-
3977743
-
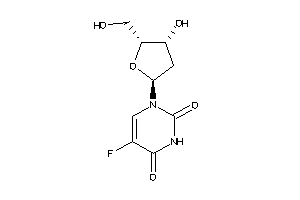
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.42 |
Binding ≤ 10μM
|
|
Z80064-6-O |
CCRF-CEM (T-cell Leukemia) (cluster #6 Of 9), Other |
Other |
500 |
0.52 |
Functional ≤ 10μM
|
|
Z80101-2-O |
COLO 320DM (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
650 |
0.51 |
Functional ≤ 10μM |
|
Z80120-3-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
92 |
0.58 |
Functional ≤ 10μM |
|
Z80164-4-O |
HT-1080 (Fibrosarcoma Cells) (cluster #4 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
45 |
0.61 |
Functional ≤ 10μM
|
|
Z80208-1-O |
L5178Y (Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
2 |
0.72 |
Functional ≤ 10μM
|
|
Z80268-2-O |
MKN-28 (Gastric Epithelial Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80269-3-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
3500 |
0.45 |
Functional ≤ 10μM |
|
Z80347-2-O |
NUGC-3 (Gastric Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
15 |
0.64 |
Functional ≤ 10μM |
|
Z80525-2-O |
SW48 (cluster #2 Of 2), Other |
Other |
130 |
0.57 |
Functional ≤ 10μM |
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80682-9-O |
A549 (Lung Carcinoma Cells) (cluster #9 Of 11), Other |
Other |
1000 |
0.49 |
Functional ≤ 10μM |
|
Z80686-1-O |
AZ-521 Cell Line (cluster #1 Of 1), Other |
Other |
50 |
0.60 |
Functional ≤ 10μM |
|
Z81048-1-O |
TSU (Prostatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
58 |
0.60 |
Functional ≤ 10μM
|
|
Z81121-1-O |
KKLS Cell Line (cluster #1 Of 1), Other |
Other |
760 |
0.50 |
Functional ≤ 10μM |
|
Z81137-1-O |
L929 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
7700 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-4-O |
LNCaP (Prostate Carcinoma) (cluster #4 Of 5), Other |
Other |
69 |
0.59 |
Functional ≤ 10μM
|
|
Z81330-1-O |
SNU- (cluster #1 Of 1), Other |
Other |
3 |
0.70 |
Functional ≤ 10μM |
|
Z81335-3-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
49 |
0.60 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.72 |
-5.12 |
-11.33 |
3 |
7 |
0 |
105 |
246.194 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.26 |
-7.87 |
-48.56 |
2 |
7 |
-1 |
108 |
245.186 |
2 |
↓
|
|
|
|
Analogs
-
3977743
-

-
3977744
-
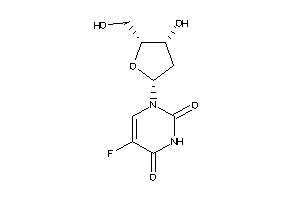
-
4045293
-
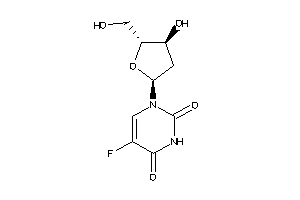
-
1457
-
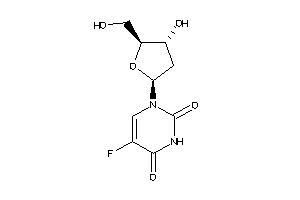
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.42 |
Binding ≤ 10μM
|
|
Z80064-6-O |
CCRF-CEM (T-cell Leukemia) (cluster #6 Of 9), Other |
Other |
500 |
0.52 |
Functional ≤ 10μM
|
|
Z80101-2-O |
COLO 320DM (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
650 |
0.51 |
Functional ≤ 10μM |
|
Z80120-3-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
92 |
0.58 |
Functional ≤ 10μM |
|
Z80164-4-O |
HT-1080 (Fibrosarcoma Cells) (cluster #4 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
45 |
0.61 |
Functional ≤ 10μM
|
|
Z80208-1-O |
L5178Y (Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
2 |
0.72 |
Functional ≤ 10μM
|
|
Z80268-2-O |
MKN-28 (Gastric Epithelial Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80269-3-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
3500 |
0.45 |
Functional ≤ 10μM |
|
Z80347-2-O |
NUGC-3 (Gastric Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
15 |
0.64 |
Functional ≤ 10μM |
|
Z80525-2-O |
SW48 (cluster #2 Of 2), Other |
Other |
130 |
0.57 |
Functional ≤ 10μM |
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80682-9-O |
A549 (Lung Carcinoma Cells) (cluster #9 Of 11), Other |
Other |
1000 |
0.49 |
Functional ≤ 10μM |
|
Z80686-1-O |
AZ-521 Cell Line (cluster #1 Of 1), Other |
Other |
50 |
0.60 |
Functional ≤ 10μM |
|
Z81048-1-O |
TSU (Prostatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
58 |
0.60 |
Functional ≤ 10μM
|
|
Z81121-1-O |
KKLS Cell Line (cluster #1 Of 1), Other |
Other |
760 |
0.50 |
Functional ≤ 10μM |
|
Z81137-1-O |
L929 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
7700 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-4-O |
LNCaP (Prostate Carcinoma) (cluster #4 Of 5), Other |
Other |
69 |
0.59 |
Functional ≤ 10μM
|
|
Z81330-1-O |
SNU- (cluster #1 Of 1), Other |
Other |
3 |
0.70 |
Functional ≤ 10μM |
|
Z81335-3-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
49 |
0.60 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.72 |
-4.03 |
-17.41 |
3 |
7 |
0 |
105 |
246.194 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.26 |
-6.78 |
-57.51 |
2 |
7 |
-1 |
108 |
245.186 |
2 |
↓
|
|
|
|
Analogs
-
3977743
-

-
3977744
-

-
4045293
-

-
1457
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.42 |
Binding ≤ 10μM
|
|
Z80064-6-O |
CCRF-CEM (T-cell Leukemia) (cluster #6 Of 9), Other |
Other |
500 |
0.52 |
Functional ≤ 10μM
|
|
Z80101-2-O |
COLO 320DM (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
650 |
0.51 |
Functional ≤ 10μM |
|
Z80120-3-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
92 |
0.58 |
Functional ≤ 10μM |
|
Z80164-4-O |
HT-1080 (Fibrosarcoma Cells) (cluster #4 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
45 |
0.61 |
Functional ≤ 10μM
|
|
Z80208-1-O |
L5178Y (Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
2 |
0.72 |
Functional ≤ 10μM
|
|
Z80268-2-O |
MKN-28 (Gastric Epithelial Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80269-3-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
3500 |
0.45 |
Functional ≤ 10μM |
|
Z80347-2-O |
NUGC-3 (Gastric Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
15 |
0.64 |
Functional ≤ 10μM |
|
Z80525-2-O |
SW48 (cluster #2 Of 2), Other |
Other |
130 |
0.57 |
Functional ≤ 10μM |
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80682-9-O |
A549 (Lung Carcinoma Cells) (cluster #9 Of 11), Other |
Other |
1000 |
0.49 |
Functional ≤ 10μM |
|
Z80686-1-O |
AZ-521 Cell Line (cluster #1 Of 1), Other |
Other |
50 |
0.60 |
Functional ≤ 10μM |
|
Z81048-1-O |
TSU (Prostatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
58 |
0.60 |
Functional ≤ 10μM
|
|
Z81121-1-O |
KKLS Cell Line (cluster #1 Of 1), Other |
Other |
760 |
0.50 |
Functional ≤ 10μM |
|
Z81137-1-O |
L929 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
7700 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-4-O |
LNCaP (Prostate Carcinoma) (cluster #4 Of 5), Other |
Other |
69 |
0.59 |
Functional ≤ 10μM
|
|
Z81330-1-O |
SNU- (cluster #1 Of 1), Other |
Other |
3 |
0.70 |
Functional ≤ 10μM |
|
Z81335-3-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
49 |
0.60 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.72 |
-4.23 |
-22.26 |
3 |
7 |
0 |
105 |
246.194 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.26 |
-6.98 |
-63.95 |
2 |
7 |
-1 |
108 |
245.186 |
2 |
↓
|
|
|
|
Analogs
-
3977743
-

-
3977744
-

-
4045293
-

-
1457
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.42 |
Binding ≤ 10μM
|
|
Z80064-6-O |
CCRF-CEM (T-cell Leukemia) (cluster #6 Of 9), Other |
Other |
500 |
0.52 |
Functional ≤ 10μM
|
|
Z80101-2-O |
COLO 320DM (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
650 |
0.51 |
Functional ≤ 10μM |
|
Z80120-3-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
92 |
0.58 |
Functional ≤ 10μM |
|
Z80164-4-O |
HT-1080 (Fibrosarcoma Cells) (cluster #4 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
45 |
0.61 |
Functional ≤ 10μM
|
|
Z80208-1-O |
L5178Y (Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
2 |
0.72 |
Functional ≤ 10μM
|
|
Z80268-2-O |
MKN-28 (Gastric Epithelial Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80269-3-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
3500 |
0.45 |
Functional ≤ 10μM |
|
Z80347-2-O |
NUGC-3 (Gastric Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
15 |
0.64 |
Functional ≤ 10μM |
|
Z80525-2-O |
SW48 (cluster #2 Of 2), Other |
Other |
130 |
0.57 |
Functional ≤ 10μM |
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
120 |
0.57 |
Functional ≤ 10μM |
|
Z80682-9-O |
A549 (Lung Carcinoma Cells) (cluster #9 Of 11), Other |
Other |
1000 |
0.49 |
Functional ≤ 10μM |
|
Z80686-1-O |
AZ-521 Cell Line (cluster #1 Of 1), Other |
Other |
50 |
0.60 |
Functional ≤ 10μM |
|
Z81048-1-O |
TSU (Prostatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
58 |
0.60 |
Functional ≤ 10μM
|
|
Z81121-1-O |
KKLS Cell Line (cluster #1 Of 1), Other |
Other |
760 |
0.50 |
Functional ≤ 10μM |
|
Z81137-1-O |
L929 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
7700 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-4-O |
LNCaP (Prostate Carcinoma) (cluster #4 Of 5), Other |
Other |
69 |
0.59 |
Functional ≤ 10μM
|
|
Z81330-1-O |
SNU- (cluster #1 Of 1), Other |
Other |
3 |
0.70 |
Functional ≤ 10μM |
|
Z81335-3-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
49 |
0.60 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.72 |
-4.89 |
-10.74 |
3 |
7 |
0 |
105 |
246.194 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.26 |
-7.64 |
-47.75 |
2 |
7 |
-1 |
108 |
245.186 |
2 |
↓
|
|
|
|
Analogs
-
1529898
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
20 |
0.51 |
Binding ≤ 10μM
|
|
THYX-1-B |
Thymidylate Synthase ThyX (cluster #1 Of 1), Bacterial |
Bacteria |
290 |
0.44 |
Binding ≤ 10μM |
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
570 |
0.42 |
Binding ≤ 10μM |
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
-5.34 |
-150.06 |
2 |
10 |
-2 |
156 |
324.157 |
4 |
↓
|
|
|
|
Analogs
-
3977743
-

-
3977744
-

-
4045293
-

-
1457
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 47 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.74 |
Binding ≤ 10μM
|
|
Z50472-2-O |
Toxoplasma Gondii (cluster #2 Of 4), Other |
Other |
960 |
0.50 |
Functional ≤ 10μM
|
|
Z80005-1-O |
3T3 (Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
600 |
0.51 |
Functional ≤ 10μM
|
|
Z80064-6-O |
CCRF-CEM (T-cell Leukemia) (cluster #6 Of 9), Other |
Other |
17 |
0.64 |
Functional ≤ 10μM
|
|
Z80136-1-O |
FM3A (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
9 |
0.66 |
Functional ≤ 10μM
|
|
Z80156-6-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #6 Of 12), Other |
Other |
12 |
0.65 |
Functional ≤ 10μM
|
|
Z80164-4-O |
HT-1080 (Fibrosarcoma Cells) (cluster #4 Of 6), Other |
Other |
180 |
0.56 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
5 |
0.68 |
Functional ≤ 10μM
|
|
Z80208-1-O |
L5178Y (Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
9 |
0.66 |
Functional ≤ 10μM
|
|
Z80682-9-O |
A549 (Lung Carcinoma Cells) (cluster #9 Of 11), Other |
Other |
9740 |
0.41 |
Functional ≤ 10μM
|
|
Z81247-7-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #7 Of 9), Other |
Other |
8500 |
0.42 |
Functional ≤ 10μM
|
|
Z81252-9-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #9 Of 11), Other |
Other |
210 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.72 |
-4.38 |
-16.24 |
3 |
7 |
0 |
105 |
246.194 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.26 |
-7.12 |
-54.96 |
2 |
7 |
-1 |
108 |
245.186 |
2 |
↓
|
|