|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.31 |
Binding ≤ 10μM
|
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.32 |
12.42 |
-98.79 |
3 |
5 |
2 |
47 |
452.426 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
67 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.92 |
13.81 |
-101.52 |
4 |
7 |
2 |
78 |
562.661 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.92 |
12.2 |
-37.03 |
3 |
7 |
1 |
77 |
561.653 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.92 |
11.82 |
-18.86 |
2 |
7 |
0 |
76 |
560.645 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
41 |
0.33 |
Binding ≤ 10μM
|
|
UTS2-1-E |
Urotensin-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.38 |
Binding ≤ 10μM
|
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
8.67 |
-83.18 |
5 |
6 |
2 |
80 |
420.557 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.96 |
5.02 |
-20.94 |
3 |
6 |
0 |
81 |
418.541 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.96 |
8.19 |
-89.57 |
5 |
6 |
2 |
84 |
420.557 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.26 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.57 |
15.04 |
-46.06 |
1 |
4 |
1 |
28 |
461.457 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.57 |
12.2 |
-10.13 |
0 |
4 |
0 |
27 |
460.449 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.57 |
15.05 |
-43.07 |
1 |
4 |
1 |
28 |
461.457 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.57 |
12.6 |
-12.35 |
0 |
4 |
0 |
27 |
460.449 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
16 |
0.35 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.25 |
Functional ≤ 10μM
|
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
750 |
0.28 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1400 |
0.26 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.57 |
14.73 |
-27.78 |
1 |
4 |
1 |
28 |
461.457 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.57 |
13.88 |
-10.16 |
0 |
4 |
0 |
27 |
460.449 |
6 |
↓
|
|
|
|
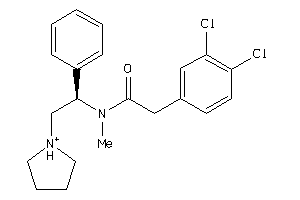
Analogs
-
3995703
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
24 |
0.41 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
600 |
0.34 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.44 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.43 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
53 |
0.39 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
150 |
0.37 |
Binding ≤ 10μM
|
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.29 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
26 |
0.41 |
ADME/T ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50594-6-O |
Mus Musculus (cluster #6 Of 9), Other |
Other |
3 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.77 |
13.65 |
-44.2 |
1 |
3 |
1 |
25 |
392.35 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
38 |
0.34 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
140 |
0.31 |
Binding ≤ 10μM
|
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.23 |
Binding ≤ 10μM
|
|
5HT2B-3-E |
Serotonin 2b (5-HT2b) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.31 |
Binding ≤ 10μM
|
|
5HT2C-3-E |
Serotonin 2c (5-HT2c) Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
85 |
0.32 |
Binding ≤ 10μM
|
|
5HT6R-2-E |
Serotonin 6 (5-HT6) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.24 |
Binding ≤ 10μM
|
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.38 |
Binding ≤ 10μM
|
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.38 |
Functional ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
ADME/T ≤ 10μM
|
|
CP343-1-E |
Cytochrome P450 3A43 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.26 |
ADME/T ≤ 10μM
|
|
DRD2-17-E |
Dopamine D2 Receptor (cluster #17 Of 24), Eukaryotic |
Eukaryotes |
100 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.85 |
11.14 |
-110.9 |
4 |
5 |
2 |
59 |
529.306 |
6 |
↓
|
|
|
|
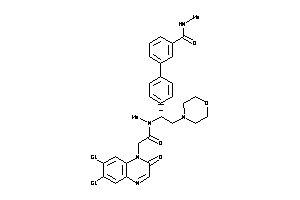
Analogs
-
42834388
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.28 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.19 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5000 |
0.18 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.28 |
9.81 |
-24.5 |
1 |
9 |
0 |
97 |
608.526 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.28 |
12.33 |
-58.19 |
2 |
9 |
1 |
98 |
609.534 |
8 |
↓
|
|
|
|
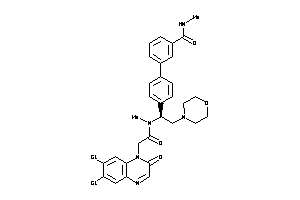
Analogs
-
42834386
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.28 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.19 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5000 |
0.18 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.28 |
10.51 |
-22.17 |
1 |
9 |
0 |
97 |
608.526 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.28 |
13.07 |
-64.28 |
2 |
9 |
1 |
98 |
609.534 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.35 |
Binding ≤ 10μM
|
|
UTS2-1-E |
Urotensin-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
4.12 |
4.72 |
-39.47 |
0 |
7 |
-1 |
79 |
538.362 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.29 |
Binding ≤ 10μM
|
|
UR2R-1-E |
Urotensin II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
11.35 |
-54.1 |
2 |
9 |
1 |
93 |
599.799 |
12 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.70 |
9.15 |
-16.11 |
1 |
9 |
0 |
91 |
598.791 |
12 |
↓
|
|