|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
4.05 |
-5.7 |
1 |
5 |
0 |
52 |
278.743 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.52 |
6.41 |
-40.72 |
2 |
5 |
1 |
53 |
279.751 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
8.11 |
-13.19 |
1 |
4 |
0 |
51 |
342.395 |
5 |
↓
|
|
|
|
Analogs
-
2000919
-
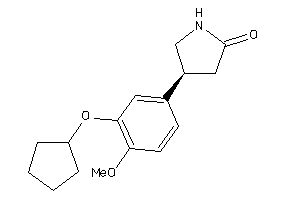
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
O77823-1-E |
Phosphodiesterase 4A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.43 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
740 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.52 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.42 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3690 |
0.38 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
622 |
0.43 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.42 |
Binding ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
720 |
0.43 |
Binding ≤ 10μM
|
|
Q8VBU5-1-E |
Phosphodiesterase 4B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
625 |
0.43 |
Functional ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.36 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 1), Other |
Other |
150 |
0.48 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
7 |
0.57 |
Binding ≤ 10μM |
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
880 |
0.42 |
Functional ≤ 10μM
|
|
Z100489-1-O |
Eosinophils (Eosinophils) (cluster #1 Of 1), Other |
Other |
6500 |
0.36 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
340 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
660 |
0.43 |
Functional ≤ 10μM |
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
4 |
0.59 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
40 |
0.52 |
Functional ≤ 10μM
|
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
1230 |
0.41 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
5.43 |
-11.26 |
1 |
4 |
0 |
48 |
275.348 |
4 |
↓
|
|
|
|
Analogs
-
4982
-
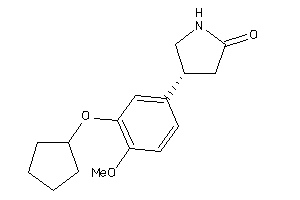
Draw
Identity
99%
90%
80%
70%
Vendors
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
O77823-1-E |
Phosphodiesterase 4A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.43 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
740 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.42 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.42 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
720 |
0.43 |
Binding ≤ 10μM
|
|
Q8VBU5-1-E |
Phosphodiesterase 4B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
625 |
0.43 |
Functional ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.36 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 1), Other |
Other |
150 |
0.48 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
7 |
0.57 |
Binding ≤ 10μM |
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
880 |
0.42 |
Functional ≤ 10μM
|
|
Z100489-1-O |
Eosinophils (Eosinophils) (cluster #1 Of 1), Other |
Other |
6500 |
0.36 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
340 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
660 |
0.43 |
Functional ≤ 10μM |
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
4 |
0.59 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
40 |
0.52 |
Functional ≤ 10μM
|
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
1230 |
0.41 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
5.44 |
-11.23 |
1 |
4 |
0 |
48 |
275.348 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.85 |
2.11 |
-7.58 |
1 |
4 |
0 |
51 |
275.348 |
4 |
↓
|
|