|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
Pyridine, 1,2,3,6-tetrahydro-1-methyl-5-(2-propyl-2H-1,2,3-triazol-4-yl)- (9CI)
Pyridine, 1,2,3,6-tetrahydro-1-m…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
15 |
0.73 |
Binding ≤ 10μM
|
|
Z104303-3-O |
Muscarinic Acetylcholine Receptor (cluster #3 Of 7), Other |
Other |
47 |
0.68 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.36 |
6.05 |
-40.87 |
1 |
4 |
1 |
35 |
207.301 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
36 |
0.74 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.55 |
Functional ≤ 10μM
|
|
ACM3-3-E |
Muscarinic Acetylcholine Receptor M3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.55 |
Functional ≤ 10μM
|
|
Z104303-3-O |
Muscarinic Acetylcholine Receptor (cluster #3 Of 7), Other |
Other |
9 |
0.80 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.19 |
1.76 |
-38.47 |
1 |
5 |
1 |
48 |
194.262 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
270 |
0.57 |
Binding ≤ 10μM
|
|
Z104303-3-O |
Muscarinic Acetylcholine Receptor (cluster #3 Of 7), Other |
Other |
300 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.87 |
6.07 |
-35.55 |
1 |
5 |
1 |
48 |
222.316 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104303-3-O |
Muscarinic Acetylcholine Receptor (cluster #3 Of 7), Other |
Other |
3290 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.28 |
4.09 |
-41.45 |
2 |
3 |
1 |
43 |
165.216 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104303-3-O |
Muscarinic Acetylcholine Receptor (cluster #3 Of 7), Other |
Other |
110 |
0.89 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
2.29 |
-4.28 |
0 |
3 |
0 |
25 |
154.213 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
33 |
1.05 |
Binding ≤ 10μM |
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
33 |
1.05 |
Binding ≤ 10μM |
|
ACHA4-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
33 |
1.05 |
Binding ≤ 10μM |
|
Z104303-3-O |
Muscarinic Acetylcholine Receptor (cluster #3 Of 7), Other |
Other |
4380 |
0.75 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.48 |
1.65 |
-41.72 |
1 |
2 |
1 |
21 |
140.206 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
48 |
1.02 |
Binding ≤ 10μM |
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
48 |
1.02 |
Binding ≤ 10μM |
|
ACHA4-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
48 |
1.02 |
Binding ≤ 10μM |
|
Z104303-3-O |
Muscarinic Acetylcholine Receptor (cluster #3 Of 7), Other |
Other |
5425 |
0.74 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.48 |
1.7 |
-41.47 |
1 |
2 |
1 |
21 |
140.206 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104303-3-O |
Muscarinic Acetylcholine Receptor (cluster #3 Of 7), Other |
Other |
110 |
0.89 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
2.18 |
-5.81 |
0 |
3 |
0 |
25 |
154.213 |
2 |
↓
|
|
|
|
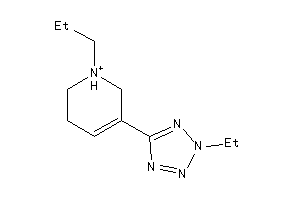
Analogs
-
13739803
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
140 |
0.64 |
Binding ≤ 10μM
|
|
Z104303-3-O |
Muscarinic Acetylcholine Receptor (cluster #3 Of 7), Other |
Other |
78 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.57 |
5.57 |
-37.63 |
1 |
5 |
1 |
48 |
208.289 |
3 |
↓
|
|
|
|
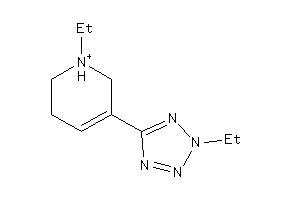
Analogs
-
13739802
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
900 |
0.53 |
Binding ≤ 10μM
|
|
Z104303-3-O |
Muscarinic Acetylcholine Receptor (cluster #3 Of 7), Other |
Other |
450 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.07 |
6.31 |
-38.16 |
1 |
5 |
1 |
48 |
222.316 |
4 |
↓
|
|