|
|
Analogs
-
120294
-
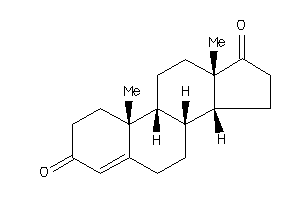
-
2046798
-
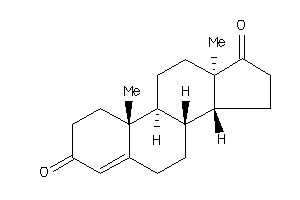
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
37 |
0.45 |
Binding ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
FABPL-1-E |
Fatty Acid-binding Protein, Liver (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.46 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
31 |
0.46 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.48 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.34 |
Binding ≤ 10μM
|
|
SGMR1-2-E |
Sigma Opioid Receptor (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
260 |
0.40 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
37 |
0.45 |
Functional ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.30 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.30 |
Functional ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2770 |
0.34 |
Functional ≤ 10μM
|
|
MCR-2-E |
Mineralocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Functional ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Functional ≤ 10μM
|
|
CP2C9-2-E |
Cytochrome P450 2C9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5500 |
0.32 |
ADME/T ≤ 10μM
|
|
ERG2-1-F |
C-8 Sterol Isomerase (cluster #1 Of 2), Fungal |
Fungi |
4430 |
0.33 |
Binding ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
6310 |
0.32 |
Functional ≤ 10μM
|
|
Z80712-1-O |
T47D (Breast Carcinoma Cells) (cluster #1 Of 7), Other |
Other |
1 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
10.64 |
-9.9 |
0 |
2 |
0 |
34 |
314.469 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
10000 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
6.3 |
-16.6 |
2 |
4 |
0 |
75 |
346.467 |
2 |
↓
|
|
|
|
Analogs
-
3814382
-
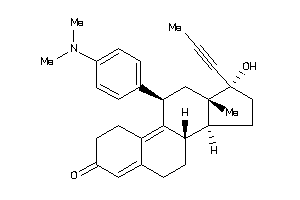
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
89 |
0.31 |
Binding ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.23 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2876 |
0.24 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
410 |
0.28 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
640 |
0.27 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.37 |
Binding ≤ 10μM |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.35 |
Functional ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2244 |
0.25 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
812 |
0.27 |
Functional ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.38 |
Functional ≤ 10μM
|
|
MCR-2-E |
Mineralocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1155 |
0.26 |
Functional ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.37 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4700 |
0.23 |
ADME/T ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
6310 |
0.23 |
Functional ≤ 10μM
|
|
Z80110-2-O |
CV-1 (Kidney Cells) (cluster #2 Of 2), Other |
Other |
10 |
0.35 |
Functional ≤ 10μM
|
|
Z80491-2-O |
SK-N-MC (Neuroepithelioma Cells) (cluster #2 Of 4), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
2 |
0.38 |
Functional ≤ 10μM
|
|
VP16-1-V |
Alpha Trans-inducing Protein (VP16) (cluster #1 Of 1), Viral |
Viruses |
1 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.60 |
1.26 |
-10.46 |
1 |
3 |
0 |
40 |
429.604 |
2 |
↓
|
|
|
|
Analogs
-
31168358
-
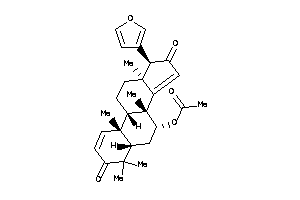
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
3400 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.23 |
12.51 |
-15.89 |
0 |
5 |
0 |
74 |
450.575 |
3 |
↓
|
|
|
|
Analogs
-
4073970
-
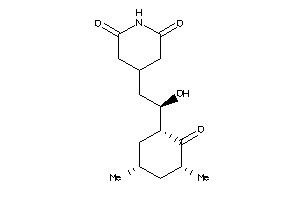
-
4536598
-
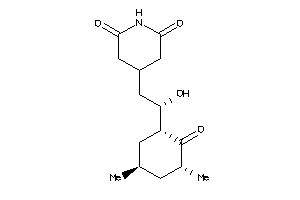
-
8579261
-
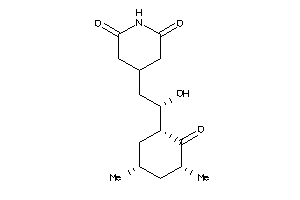
-
11592912
-
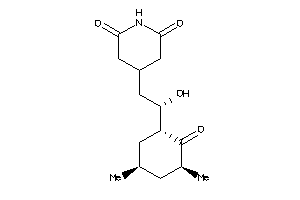
-
11592913
-
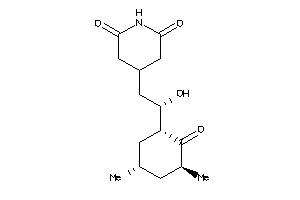
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.38 |
Binding ≤ 10μM
|
|
Z50347-2-O |
Saccharomyces Cerevisiae (cluster #2 Of 3), Other |
Other |
88 |
0.49 |
Functional ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
200 |
0.47 |
Functional ≤ 10μM
|
|
Z50466-5-O |
Trypanosoma Cruzi (cluster #5 Of 8), Other |
Other |
400 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
930 |
0.42 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
5000 |
0.37 |
Functional ≤ 10μM
|
|
Z80026-2-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
3600 |
0.38 |
Functional ≤ 10μM |
|
Z80224-6-O |
MCF7 (Breast Carcinoma Cells) (cluster #6 Of 14), Other |
Other |
1500 |
0.41 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
3500 |
0.38 |
Functional ≤ 10μM
|
|
Z80485-2-O |
SK-MEL-28 (Melanoma Cells) (cluster #2 Of 6), Other |
Other |
1000 |
0.42 |
Functional ≤ 10μM
|
|
Z80490-1-O |
SK-MES-1 (cluster #1 Of 4), Other |
Other |
2700 |
0.39 |
Functional ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 7), Other |
Other |
530 |
0.44 |
Functional ≤ 10μM
|
|
Z80653-1-O |
9L (Glioma Cells) (cluster #1 Of 2), Other |
Other |
200 |
0.47 |
Functional ≤ 10μM
|
|
Z80874-4-O |
CEM (T-cell Leukemia) (cluster #4 Of 7), Other |
Other |
80 |
0.50 |
Functional ≤ 10μM
|
|
Z81020-4-O |
HepG2 (Hepatoblastoma Cells) (cluster #4 Of 8), Other |
Other |
2500 |
0.39 |
Functional ≤ 10μM
|
|
Z81072-7-O |
Jurkat (Acute Leukemic T-cells) (cluster #7 Of 10), Other |
Other |
930 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-3-O |
LNCaP (Prostate Carcinoma) (cluster #3 Of 5), Other |
Other |
1200 |
0.41 |
Functional ≤ 10μM
|
|
Z81247-6-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #6 Of 9), Other |
Other |
533 |
0.44 |
Functional ≤ 10μM
|
|
Z81252-4-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #4 Of 11), Other |
Other |
1200 |
0.41 |
Functional ≤ 10μM |
|
Z80005-2-O |
3T3 (Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
912 |
0.42 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
3.22 |
-17 |
2 |
5 |
0 |
83 |
281.352 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
794 |
0.19 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.25 |
19.64 |
-18.09 |
0 |
8 |
0 |
73 |
610.847 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.25 |
15.86 |
-34 |
1 |
8 |
1 |
74 |
611.855 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.96 |
11.84 |
-46.11 |
0 |
3 |
-1 |
57 |
365.836 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
5.55 |
9.3 |
-8.83 |
2 |
3 |
0 |
58 |
366.844 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.55 |
10.22 |
-45.56 |
1 |
3 |
-1 |
60 |
365.836 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTN1-3-E |
Protein-tyrosine Phosphatase 1B (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5930 |
0.22 |
Binding ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
8600 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.81 |
9.73 |
-53.6 |
2 |
4 |
-1 |
81 |
471.702 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.81 |
7.75 |
-7.1 |
3 |
4 |
0 |
78 |
472.71 |
1 |
↓
|
|
|
|
Analogs
-
3800039
-
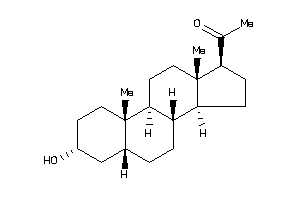
Draw
Identity
99%
90%
80%
70%
Vendors
-
-
- SHG0000527
- SHG0000071
- SHG0000774
-
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA2-4-E |
GABA Receptor Alpha-2 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA3-4-E |
GABA Receptor Alpha-3 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA4-4-E |
GABA Receptor Alpha-4 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA5-2-E |
GABA Receptor Alpha-5 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA6-3-E |
GABA Receptor Alpha-6 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1-2-E |
GABA Receptor Beta-1 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRD-3-E |
GABA Receptor Delta Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRE-3-E |
GABA Receptor Epsilon Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRG1-2-E |
GABA Receptor Gamma-1 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRG3-3-E |
GABA Receptor Gamma-3 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRP-3-E |
GABA Receptor Pi Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
Q91ZM7-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA1-2-E |
GABA Receptor Alpha-1 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
160 |
0.41 |
Functional ≤ 10μM
|
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.41 |
Functional ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.41 |
Functional ≤ 10μM
|
|
Z104301-2-O |
GABA-A Receptor; Anion Channel (cluster #2 Of 8), Other |
Other |
91 |
0.43 |
Binding ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
10000 |
0.30 |
Functional ≤ 10μM
|
|
Z50573-2-O |
Xenopus Laevis (cluster #2 Of 2), Other |
Other |
5500 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
8.25 |
-5.76 |
1 |
2 |
0 |
37 |
318.501 |
1 |
↓
|
|
|
|
Analogs
-
4716552
-
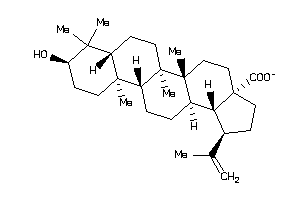
-
4995154
-
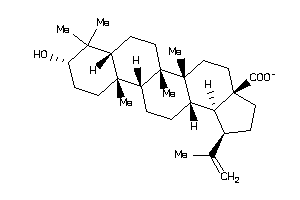
-
4995155
-
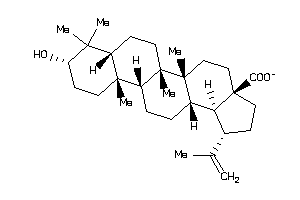
-
4995156
-
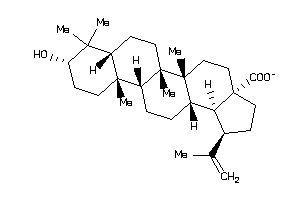
-
8951991
-
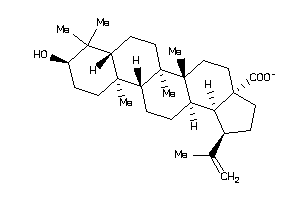
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPOLB-2-E |
DNA Polymerase Beta (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6500 |
0.22 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2170 |
0.24 |
Functional ≤ 10μM
|
|
NR1H4-2-E |
Bile Acid Receptor FXR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z103202-1-O |
8505C (cluster #1 Of 2), Other |
Other |
7260 |
0.22 |
Functional ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
5200 |
0.22 |
Functional ≤ 10μM
|
|
Z50459-6-O |
Leishmania Donovani (cluster #6 Of 8), Other |
Other |
4100 |
0.23 |
Functional ≤ 10μM
|
|
Z50602-3-O |
Human Herpesvirus 1 (cluster #3 Of 5), Other |
Other |
8200 |
0.22 |
Functional ≤ 10μM
|
|
Z50607-8-O |
Human Immunodeficiency Virus 1 (cluster #8 Of 10), Other |
Other |
3100 |
0.23 |
Functional ≤ 10μM
|
|
Z50636-1-O |
Sindbis Virus (cluster #1 Of 3), Other |
Other |
500 |
0.27 |
Functional ≤ 10μM
|
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
7000 |
0.22 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
6480 |
0.22 |
Functional ≤ 10μM
|
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
6650 |
0.22 |
Functional ≤ 10μM
|
|
Z80897-2-O |
H9 (T-lymphoid Cells) (cluster #2 Of 2), Other |
Other |
1400 |
0.25 |
Functional ≤ 10μM
|
|
Z81020-2-O |
HepG2 (Hepatoblastoma Cells) (cluster #2 Of 8), Other |
Other |
36 |
0.32 |
Functional ≤ 10μM
|
|
Z81072-2-O |
Jurkat (Acute Leukemic T-cells) (cluster #2 Of 10), Other |
Other |
27 |
0.32 |
Functional ≤ 10μM
|
|
Z81247-4-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #4 Of 9), Other |
Other |
26 |
0.32 |
Functional ≤ 10μM
|
|
Z80897-2-O |
H9 (T-lymphoid Cells) (cluster #2 Of 2), Other |
Other |
900 |
0.26 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.04 |
12.1 |
-48.88 |
1 |
3 |
-1 |
60 |
455.703 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.04 |
10.34 |
-5.48 |
2 |
3 |
0 |
58 |
456.711 |
2 |
↓
|
|
|
|
Analogs
-
120294
-

-
2046798
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
9.29 |
-10.68 |
0 |
2 |
0 |
34 |
286.415 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
3.06 |
9.39 |
-9.03 |
0 |
2 |
0 |
34 |
286.415 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
7.90 |
18.51 |
-6.65 |
0 |
4 |
0 |
68 |
536.797 |
11 |
↓
|
|
|
|
Analogs
-
59725637
-
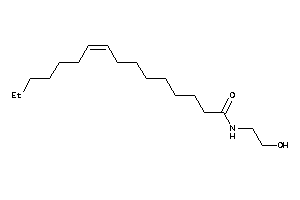
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-3-E |
Cannabinoid CB1 Receptor (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1100 |
0.36 |
Binding ≤ 10μM
|
|
PPARA-2-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
110 |
0.42 |
Binding ≤ 10μM
|
|
TRPV1-3-E |
Vanilloid Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
890 |
0.37 |
Functional ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
3162 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.81 |
-1.63 |
-8.83 |
2 |
3 |
0 |
49 |
325.537 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.69 |
1.88 |
-12.03 |
1 |
3 |
0 |
54 |
326.436 |
6 |
↓
|
|
|
|
Analogs
-
17005417
-
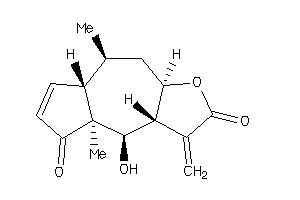
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.34 |
4.46 |
-11.54 |
1 |
4 |
0 |
64 |
262.305 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
10.37 |
-11.66 |
0 |
2 |
0 |
34 |
312.453 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
3.62 |
10.62 |
-10.79 |
0 |
2 |
0 |
34 |
312.453 |
0 |
↓
|
|
|
|
Analogs
-
5157637
-
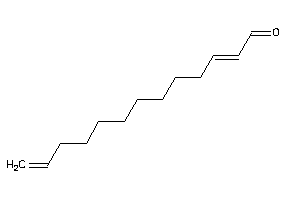
-
44712396
-
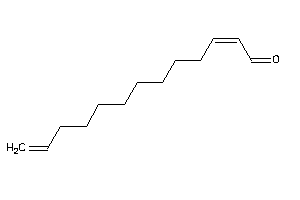
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
390 |
0.90 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
6.5 |
-5.96 |
0 |
1 |
0 |
17 |
140.226 |
6 |
↓
|
|
|
|
Analogs
-
4947897
-
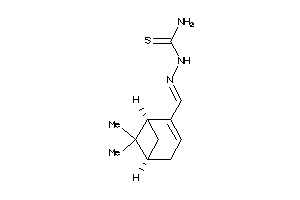
-
36502465
-
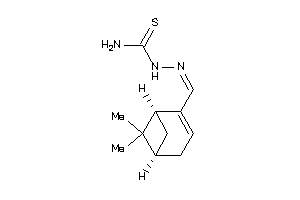
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
7200 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
5.98 |
-9.38 |
3 |
3 |
0 |
50 |
223.345 |
3 |
↓
|
|
|
|
Analogs
-
36502465
-

-
3229106
-
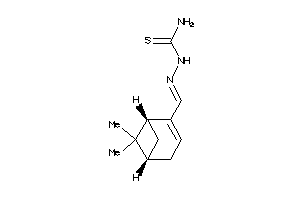
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
7200 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
-2.8 |
-9.36 |
3 |
3 |
0 |
50 |
223.345 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1aR-(1aalpha,1bbeta,4abeta,7aalpha,7balpha,8alpha,9beta,9aalpha))-Tetradecanoic acid, 9a-(acetyloxy)-1a,1b,4,4a,5,7a,7b,8,9,9a-decahydro-7b-hydroxy-3-(hydroxymethyl)-4a-methoxy-1,1,6,8-tetramethyl-1H-cyclopropa(3,4)benz(1,2-e)azulen-9-yl ester; 4-O-Methy
(1aR-(1aalpha,1bbeta,4abeta,7aal…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
6310 |
0.16 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.42 |
15.58 |
-11.76 |
2 |
8 |
0 |
119 |
630.863 |
18 |
↓
|
|
|
|
|
|
|
Analogs
-
1531550
-
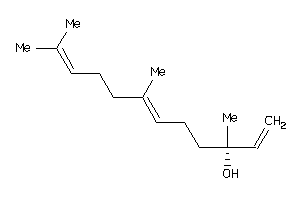
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
120 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.06 |
7.92 |
-2.02 |
1 |
1 |
0 |
20 |
222.372 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
8623412
-
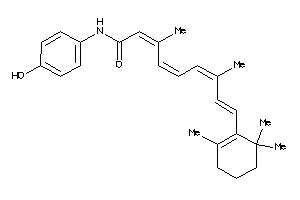
-
9228221
-
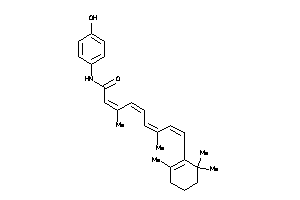
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RET1-1-E |
Cellular Retinol-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.33 |
Binding ≤ 10μM
|
|
RET2-1-E |
Cellular Retinol-binding Protein II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.33 |
Binding ≤ 10μM
|
|
RET3-1-E |
Interstitial Retinol-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.33 |
Binding ≤ 10μM
|
|
RET4-1-E |
Plasma Retinol-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.33 |
Binding ≤ 10μM
|
|
RET5-1-E |
Cellular Retinol-binding Protein III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.33 |
Binding ≤ 10μM
|
|
RET7-1-E |
Cellular Retinol-binding Protein IV (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.33 |
Binding ≤ 10μM
|
|
RET4-1-E |
Plasma Retinol-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2372 |
0.27 |
Functional ≤ 10μM |
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
7943 |
0.25 |
Functional ≤ 10μM
|
|
Z80224-2-O |
MCF7 (Breast Carcinoma Cells) (cluster #2 Of 14), Other |
Other |
150 |
0.33 |
Functional ≤ 10μM
|
|
Z80390-8-O |
PC-3 (Prostate Carcinoma Cells) (cluster #8 Of 10), Other |
Other |
3600 |
0.26 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
7500 |
0.25 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
7700 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.88 |
11.69 |
-8.39 |
2 |
3 |
0 |
49 |
391.555 |
6 |
↓
|
|