|
|
Analogs
-
5161573
-
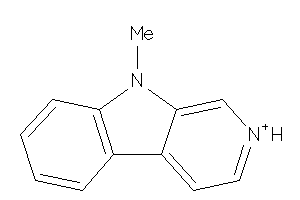
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
541 |
0.63 |
Binding ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
340 |
0.65 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
450 |
0.63 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.05 |
0.8 |
-30.36 |
1 |
2 |
1 |
19 |
183.234 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100497-1-O |
Bovine Viral Diarrhea Virus (cluster #1 Of 3), Other |
Other |
4300 |
0.54 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
2400 |
0.56 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.05 |
0.8 |
-29.95 |
1 |
2 |
1 |
19 |
183.234 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
4500 |
0.27 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.33 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
ADO-1-E |
Aldehyde Oxidase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
33 |
0.37 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
8 |
0.40 |
Functional ≤ 10μM
|
|
Z80578-1-O |
UV4 (cluster #1 Of 1), Other |
Other |
140 |
0.34 |
Functional ≤ 10μM
|
|
Z80667-1-O |
AA8 (cluster #1 Of 2), Other |
Other |
1700 |
0.29 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
800 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.56 |
11.66 |
-76.98 |
3 |
4 |
2 |
40 |
401.982 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 33 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
4500 |
0.27 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.33 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
ADO-1-E |
Aldehyde Oxidase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
33 |
0.37 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
8 |
0.40 |
Functional ≤ 10μM
|
|
Z80578-1-O |
UV4 (cluster #1 Of 1), Other |
Other |
140 |
0.34 |
Functional ≤ 10μM
|
|
Z80667-1-O |
AA8 (cluster #1 Of 2), Other |
Other |
1700 |
0.29 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
800 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.56 |
11.53 |
-77.73 |
3 |
4 |
2 |
40 |
401.982 |
9 |
↓
|
|
|
|
Analogs
-
1530861
-
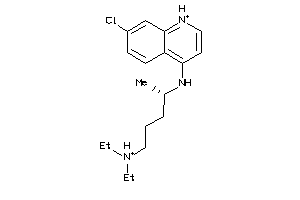
-
5850156
-
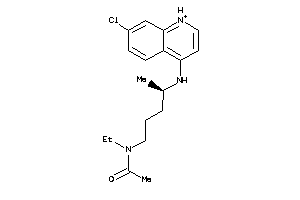
-
5850183
-
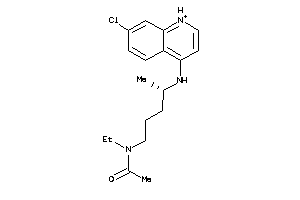
-
6424763
-
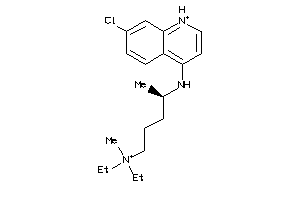
-
19144226
-
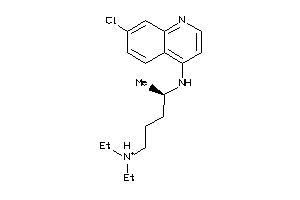
Draw
Identity
99%
90%
80%
70%
Vendors
And 66 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.41 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2512 |
0.36 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.34 |
Binding ≤ 10μM
|
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
400 |
0.41 |
Binding ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100498-2-O |
Hepatocytes (cluster #2 Of 2), Other |
Other |
1900 |
0.36 |
Functional ≤ 10μM
|
|
Z101682-1-O |
BK Polyomavirus (cluster #1 Of 2), Other |
Other |
1700 |
0.37 |
Functional ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
14 |
0.50 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
90 |
0.45 |
Functional ≤ 10μM
|
|
Z50136-2-O |
Plasmodium Falciparum (isolate FcB1 / Columbia) (cluster #2 Of 3), Other |
Other |
167 |
0.43 |
Functional ≤ 10μM
|
|
Z50424-2-O |
Cryptosporidium Parvum (cluster #2 Of 2), Other |
Other |
27 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
900 |
0.38 |
Functional ≤ 10μM
|
|
Z50468-2-O |
Giardia Intestinalis (cluster #2 Of 4), Other |
Other |
401 |
0.41 |
Functional ≤ 10μM
|
|
Z50473-4-O |
Plasmodium Berghei (cluster #4 Of 5), Other |
Other |
72 |
0.45 |
Functional ≤ 10μM
|
|
Z50474-1-O |
Plasmodium Yoelii (cluster #1 Of 1), Other |
Other |
106 |
0.44 |
Functional ≤ 10μM
|
|
Z80136-1-O |
FM3A (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
40 |
0.47 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
4000 |
0.34 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
9670 |
0.32 |
Functional ≤ 10μM
|
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 2), Other |
Other |
5100 |
0.34 |
ADME/T ≤ 10μM
|
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 3), Other |
Other |
32 |
0.48 |
ADME/T ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
64 |
0.46 |
ADME/T ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 1), Other |
Other |
8300 |
0.32 |
ADME/T ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
600 |
0.40 |
ADME/T ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
7700 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
10.55 |
-81.59 |
3 |
3 |
2 |
31 |
321.896 |
8 |
↓
|
|
|
|
Analogs
-
5850156
-

-
5850183
-

-
6424763
-

-
19144226
-

-
19144231
-
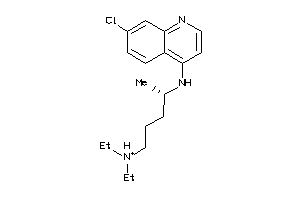
Draw
Identity
99%
90%
80%
70%
Vendors
And 64 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.41 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2512 |
0.36 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.34 |
Binding ≤ 10μM
|
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
400 |
0.41 |
Binding ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100498-2-O |
Hepatocytes (cluster #2 Of 2), Other |
Other |
1900 |
0.36 |
Functional ≤ 10μM
|
|
Z101682-1-O |
BK Polyomavirus (cluster #1 Of 2), Other |
Other |
1700 |
0.37 |
Functional ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
14 |
0.50 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
90 |
0.45 |
Functional ≤ 10μM
|
|
Z50136-2-O |
Plasmodium Falciparum (isolate FcB1 / Columbia) (cluster #2 Of 3), Other |
Other |
167 |
0.43 |
Functional ≤ 10μM
|
|
Z50424-2-O |
Cryptosporidium Parvum (cluster #2 Of 2), Other |
Other |
27 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
900 |
0.38 |
Functional ≤ 10μM
|
|
Z50468-2-O |
Giardia Intestinalis (cluster #2 Of 4), Other |
Other |
401 |
0.41 |
Functional ≤ 10μM
|
|
Z50473-4-O |
Plasmodium Berghei (cluster #4 Of 5), Other |
Other |
72 |
0.45 |
Functional ≤ 10μM
|
|
Z50474-1-O |
Plasmodium Yoelii (cluster #1 Of 1), Other |
Other |
106 |
0.44 |
Functional ≤ 10μM
|
|
Z80136-1-O |
FM3A (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
40 |
0.47 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
4000 |
0.34 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
9670 |
0.32 |
Functional ≤ 10μM
|
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 2), Other |
Other |
5100 |
0.34 |
ADME/T ≤ 10μM
|
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 3), Other |
Other |
32 |
0.48 |
ADME/T ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
64 |
0.46 |
ADME/T ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 1), Other |
Other |
8300 |
0.32 |
ADME/T ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
600 |
0.40 |
ADME/T ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
7700 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
10.55 |
-81.48 |
3 |
3 |
2 |
31 |
321.896 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
400 |
0.34 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
2750 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.55 |
1.5 |
-35.15 |
0 |
5 |
1 |
40 |
348.378 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-1-O |
Trypanosoma Brucei (cluster #1 Of 6), Other |
Other |
3000 |
0.43 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
755 |
0.48 |
Functional ≤ 10μM |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
440 |
0.49 |
Functional ≤ 10μM
|
|
Z50459-8-O |
Leishmania Donovani (cluster #8 Of 8), Other |
Other |
2680 |
0.43 |
Functional ≤ 10μM
|
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
220 |
0.52 |
Functional ≤ 10μM
|
|
Z50725-7-O |
Trypanosoma Brucei Rhodesiense (cluster #7 Of 7), Other |
Other |
600 |
0.48 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
1500 |
0.45 |
Functional ≤ 10μM
|
|
Z80695-2-O |
BE (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
1280 |
0.46 |
Functional ≤ 10μM
|
|
Z81240-1-O |
MAC15A (Colon Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
3950 |
0.42 |
Functional ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
1050 |
0.47 |
ADME/T ≤ 10μM |
|
Z81057-4-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #4 Of 5), Other |
Other |
1180 |
0.46 |
ADME/T ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
1120 |
0.46 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
0.21 |
-16.41 |
0 |
2 |
0 |
17 |
232.286 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.49 |
15.51 |
-129.88 |
5 |
8 |
3 |
78 |
686.708 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
8.49 |
12.84 |
-32.97 |
3 |
8 |
1 |
76 |
684.692 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
1000 |
0.31 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
1000 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.58 |
-4.71 |
-16.37 |
2 |
5 |
0 |
71 |
397.887 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.40 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z100501-1-O |
Lu1 (Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
80 |
0.52 |
Functional ≤ 10μM
|
|
Z103204-1-O |
A427 (cluster #1 Of 4), Other |
Other |
890 |
0.45 |
Functional ≤ 10μM
|
|
Z103205-3-O |
A431 (cluster #3 Of 4), Other |
Other |
470 |
0.47 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
501 |
0.46 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
73 |
0.53 |
Functional ≤ 10μM
|
|
Z80008-2-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
890 |
0.45 |
Functional ≤ 10μM
|
|
Z80036-2-O |
BaF3 (IL-3-dependent Pro-B-cells) (cluster #2 Of 3), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z80145-1-O |
H69 (cluster #1 Of 4), Other |
Other |
1800 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-10-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #10 Of 12), Other |
Other |
660 |
0.46 |
Functional ≤ 10μM
|
|
Z80164-3-O |
HT-1080 (Fibrosarcoma Cells) (cluster #3 Of 6), Other |
Other |
4300 |
0.40 |
Functional ≤ 10μM
|
|
Z80186-10-O |
K562 (Erythroleukemia Cells) (cluster #10 Of 11), Other |
Other |
380 |
0.47 |
Functional ≤ 10μM
|
|
Z80193-4-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #4 Of 12), Other |
Other |
500 |
0.46 |
Functional ≤ 10μM |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
430 |
0.47 |
Functional ≤ 10μM
|
|
Z80322-1-O |
NCI-H358 (Lung Carcinama Cells) (cluster #1 Of 2), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z80509-1-O |
SNU-638 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.42 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
800 |
0.45 |
Functional ≤ 10μM
|
|
Z80682-7-O |
A549 (Lung Carcinoma Cells) (cluster #7 Of 11), Other |
Other |
470 |
0.47 |
Functional ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2300 |
0.42 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
410 |
0.47 |
Functional ≤ 10μM
|
|
Z81024-8-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #8 Of 8), Other |
Other |
510 |
0.46 |
Functional ≤ 10μM
|
|
Z81115-3-O |
KB (Squamous Cell Carcinoma) (cluster #3 Of 6), Other |
Other |
812 |
0.45 |
Functional ≤ 10μM
|
|
Z81115-3-O |
KB (Squamous Cell Carcinoma) (cluster #3 Of 6), Other |
Other |
1780 |
0.42 |
Functional ≤ 10μM |
|
Z81170-5-O |
LNCaP (Prostate Carcinoma) (cluster #5 Of 5), Other |
Other |
3250 |
0.40 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
310 |
0.48 |
Functional ≤ 10μM
|
|
Z81252-10-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #10 Of 11), Other |
Other |
2300 |
0.42 |
Functional ≤ 10μM
|
|
Z81277-1-O |
NCI-N417 (cluster #1 Of 2), Other |
Other |
870 |
0.45 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 4), Other |
Other |
2600 |
0.41 |
ADME/T ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 1), Other |
Other |
5010 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
190 |
0.50 |
ADME/T ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
3820 |
0.40 |
ADME/T ≤ 10μM |
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
5500 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
2070 |
0.42 |
ADME/T ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1820 |
0.42 |
ADME/T ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 1), Other |
Other |
3 |
0.63 |
ADME/T ≤ 10μM
|
|
Z81057-4-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #4 Of 5), Other |
Other |
370 |
0.47 |
ADME/T ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
5300 |
0.39 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.28 |
8.05 |
-7.84 |
1 |
2 |
0 |
29 |
246.313 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.28 |
8.52 |
-35.14 |
2 |
2 |
1 |
30 |
247.321 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
6800 |
0.33 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
-1.86 |
-12.24 |
2 |
4 |
0 |
54 |
311.772 |
2 |
↓
|
|
|
|
Analogs
-
42455656
-
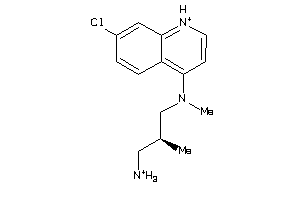
-
42455657
-
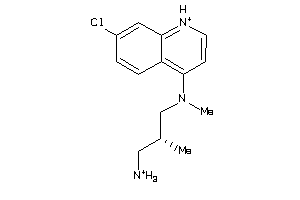
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7 |
0.57 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
23 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
-0.33 |
-85.24 |
3 |
3 |
2 |
30 |
293.842 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
51 |
0.41 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
62 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.22 |
2.82 |
-85.32 |
3 |
3 |
2 |
30 |
355.448 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
910 |
0.42 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
13 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
8.73 |
-94.78 |
3 |
3 |
2 |
31 |
291.826 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.62 |
6.06 |
-7.06 |
1 |
3 |
0 |
28 |
289.81 |
4 |
↓
|
|
|
|
Analogs
-
4115356
-
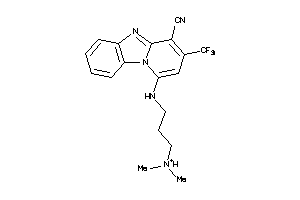
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
2080 |
0.28 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.37 |
12.26 |
-61.07 |
2 |
5 |
1 |
58 |
390.433 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
240 |
0.46 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
240 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
3.99 |
-33.91 |
2 |
4 |
1 |
39 |
292.79 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.56 |
3.6 |
-8.85 |
1 |
4 |
0 |
37 |
291.782 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.56 |
6.32 |
-98.57 |
3 |
4 |
2 |
40 |
293.798 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
1338 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
-3.59 |
-89.89 |
5 |
3 |
2 |
53 |
265.788 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
1338 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
-3.58 |
-89.86 |
5 |
3 |
2 |
53 |
265.788 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
2050 |
0.35 |
Functional ≤ 10μM |
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 2), Other |
Other |
10000 |
0.30 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
-1.96 |
-11.32 |
2 |
4 |
0 |
54 |
346.217 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
690 |
0.39 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
7.02 |
-10.6 |
2 |
4 |
0 |
54 |
295.317 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
4720 |
0.34 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
6.99 |
-12.74 |
2 |
4 |
0 |
54 |
295.317 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
9800 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
5.92 |
-27 |
2 |
2 |
1 |
30 |
183.234 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.81 |
5.53 |
-10.38 |
1 |
2 |
0 |
29 |
182.226 |
0 |
↓
|
|
|
|
Analogs
-
4674079
-
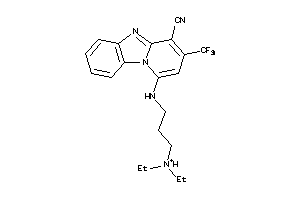
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
3160 |
0.30 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
3.55 |
-62.28 |
2 |
5 |
1 |
57 |
362.379 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
3430 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
3.42 |
-65.75 |
2 |
5 |
1 |
57 |
348.352 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
1900 |
0.30 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.10 |
3.53 |
-64.61 |
2 |
5 |
1 |
57 |
376.406 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
267 |
0.33 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
267 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.08 |
1.32 |
-84.98 |
3 |
4 |
2 |
39 |
379.548 |
10 |
↓
|
|
|
|
Analogs
-
6815
-
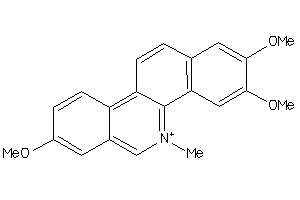
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.00 |
0.77 |
-36.44 |
1 |
5 |
1 |
51 |
350.394 |
3 |
↓
|
|
|
|
Analogs
-
20013
-
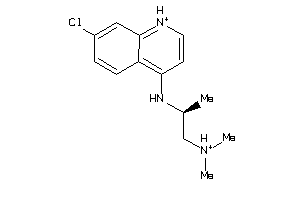
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
97 |
0.55 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
47 |
0.57 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
64 |
0.56 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.14 |
-2.4 |
-95.26 |
4 |
3 |
2 |
42 |
265.788 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.60 |
10.21 |
-76.51 |
2 |
6 |
2 |
41 |
537.539 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.60 |
9.81 |
-38.25 |
1 |
6 |
1 |
40 |
536.531 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.60 |
12.59 |
-154.37 |
3 |
6 |
3 |
42 |
538.547 |
6 |
↓
|
|
|
|
Analogs
-
36473909
-
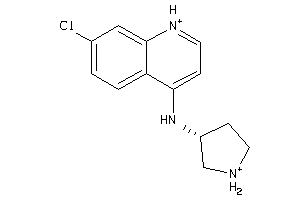
-
36473910
-
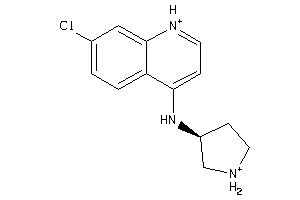
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7 |
0.60 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
13 |
0.58 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
7.88 |
-93.74 |
3 |
3 |
2 |
31 |
277.799 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.12 |
4.95 |
-7.11 |
1 |
3 |
0 |
28 |
275.783 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.12 |
7.49 |
-49.66 |
2 |
3 |
1 |
29 |
276.791 |
4 |
↓
|
|
|
|
Analogs
-
6815
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
70 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.31 |
2.72 |
-36.82 |
0 |
5 |
1 |
40 |
364.421 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
2300 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
5.34 |
-32.32 |
3 |
3 |
1 |
46 |
271.727 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.34 |
4.92 |
-7.72 |
2 |
3 |
0 |
45 |
270.719 |
2 |
↓
|
|
|
|
Analogs
-
4809430
-
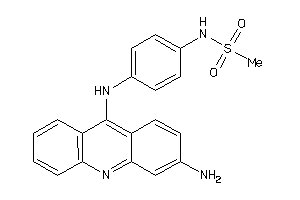
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
-8.58 |
-20.69 |
6 |
7 |
0 |
123 |
393.472 |
4 |
↓
|
|
|
|
Analogs
-
4520888
-
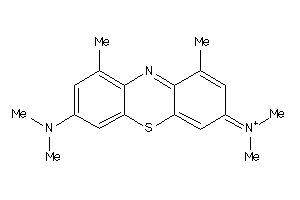
Draw
Identity
99%
90%
80%
70%
Vendors
And 46 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.09 |
8.07 |
-21.27 |
0 |
3 |
1 |
19 |
284.408 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
IGF1R-1-E |
Insulin-like Growth Factor 1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.30 |
Binding ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
390 |
0.37 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.66 |
6.69 |
-29.47 |
3 |
5 |
1 |
68 |
342.806 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.48 |
7.18 |
-28.99 |
3 |
5 |
1 |
64 |
342.806 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.48 |
6.8 |
-12.43 |
2 |
5 |
0 |
63 |
341.798 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
90 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.66 |
-0.51 |
-83.16 |
3 |
3 |
2 |
30 |
319.88 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
3 |
0.57 |
Functional ≤ 10μM
|
|
Z50725-7-O |
Trypanosoma Brucei Rhodesiense (cluster #7 Of 7), Other |
Other |
18 |
0.52 |
Functional ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
860 |
0.40 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.58 |
3.85 |
-76.3 |
9 |
5 |
2 |
119 |
279.347 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
194 |
0.63 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.39 |
7.42 |
-33.48 |
1 |
2 |
1 |
20 |
217.679 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z103205-3-O |
A431 (cluster #3 Of 4), Other |
Other |
340 |
0.50 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
440 |
0.49 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
440 |
0.49 |
Functional ≤ 10μM
|
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
1300 |
0.46 |
Functional ≤ 10μM
|
|
Z80120-4-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #4 Of 5), Other |
Other |
1440 |
0.45 |
Functional ≤ 10μM
|
|
Z80682-11-O |
A549 (Lung Carcinoma Cells) (cluster #11 Of 11), Other |
Other |
550 |
0.49 |
Functional ≤ 10μM
|
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 4), Other |
Other |
8100 |
0.40 |
Functional ≤ 10μM
|
|
Z81225-2-O |
M109 (Lung Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
1000 |
0.47 |
Functional ≤ 10μM
|
|
Z81240-1-O |
MAC15A (Colon Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
9650 |
0.39 |
Functional ≤ 10μM
|
|
Z100094-1-O |
MLF (Lung Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
4900 |
0.41 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.42 |
8.58 |
-23.57 |
1 |
2 |
1 |
20 |
233.294 |
0 |
↓
|
|