|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
-1.68 |
-9.33 |
3 |
5 |
0 |
76 |
334.354 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.34 |
-1.42 |
-81.02 |
5 |
5 |
2 |
79 |
336.37 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.34 |
-1.57 |
-38.28 |
4 |
5 |
1 |
78 |
335.362 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
1000 |
0.56 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
7.21 |
-9.17 |
0 |
4 |
0 |
59 |
200.197 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.00 |
7.42 |
-36.8 |
1 |
4 |
1 |
60 |
201.205 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z103205-3-O |
A431 (cluster #3 Of 4), Other |
Other |
340 |
0.50 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
440 |
0.49 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
440 |
0.49 |
Functional ≤ 10μM
|
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
1300 |
0.46 |
Functional ≤ 10μM
|
|
Z80120-4-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #4 Of 5), Other |
Other |
1440 |
0.45 |
Functional ≤ 10μM
|
|
Z80682-11-O |
A549 (Lung Carcinoma Cells) (cluster #11 Of 11), Other |
Other |
550 |
0.49 |
Functional ≤ 10μM
|
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 4), Other |
Other |
8100 |
0.40 |
Functional ≤ 10μM
|
|
Z81225-2-O |
M109 (Lung Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
1000 |
0.47 |
Functional ≤ 10μM
|
|
Z81240-1-O |
MAC15A (Colon Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
9650 |
0.39 |
Functional ≤ 10μM
|
|
Z100094-1-O |
MLF (Lung Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
4900 |
0.41 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.42 |
8.58 |
-23.57 |
1 |
2 |
1 |
20 |
233.294 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
6700 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.70 |
7.43 |
-8.22 |
1 |
2 |
0 |
29 |
252.704 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.70 |
7.83 |
-23.51 |
2 |
2 |
1 |
30 |
253.712 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.33 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.29 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
Binding ≤ 10μM
|
|
Q5EI73-1-E |
Protein Farnesyltransferase Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.32 |
Binding ≤ 10μM
|
|
Q5EI74-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.32 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.29 |
Functional ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.29 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4060 |
0.22 |
ADME/T ≤ 10μM
|
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
4 |
0.35 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
2 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.09 |
13.62 |
-14.28 |
2 |
5 |
0 |
66 |
489.406 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.09 |
14.08 |
-48.31 |
3 |
5 |
1 |
67 |
490.414 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.09 |
14.29 |
-131.87 |
4 |
5 |
2 |
69 |
491.422 |
4 |
↓
|
|
|
|
Analogs
-
4521067
-
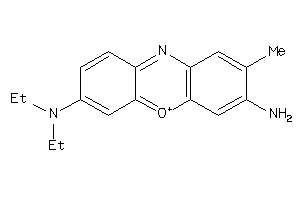
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
11.31 |
-9.05 |
0 |
4 |
0 |
31 |
324.448 |
6 |
↓
|
|
Ref
Reference (pH 7)
|
0.96 |
10.23 |
-19.87 |
0 |
4 |
1 |
32 |
324.448 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.32 |
12.63 |
-36.95 |
2 |
5 |
1 |
64 |
310.425 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.22 |
12.49 |
-16.64 |
2 |
5 |
0 |
64 |
309.417 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.41 |
12.63 |
-13.25 |
2 |
5 |
0 |
64 |
309.417 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
1.03 |
-13.88 |
0 |
2 |
0 |
29 |
183.21 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.38 |
1.15 |
-34.82 |
1 |
2 |
1 |
31 |
184.218 |
2 |
↓
|
|
|
|
Analogs
-
1530580
-
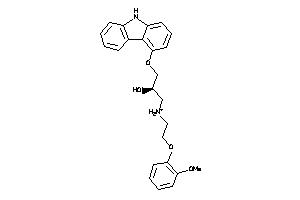
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
7.38 |
-51.26 |
4 |
6 |
1 |
80 |
407.49 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.56 |
6.05 |
-14.85 |
3 |
6 |
0 |
76 |
406.482 |
10 |
↓
|
|
|
|
Analogs
-
1530579
-
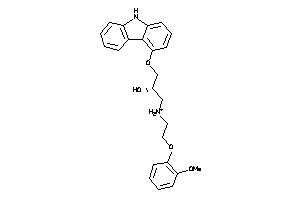
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
7.36 |
-51.12 |
4 |
6 |
1 |
80 |
407.49 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.56 |
5.94 |
-15.03 |
3 |
6 |
0 |
76 |
406.482 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-1-O |
Trypanosoma Brucei (cluster #1 Of 6), Other |
Other |
3000 |
0.43 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
755 |
0.48 |
Functional ≤ 10μM |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
440 |
0.49 |
Functional ≤ 10μM
|
|
Z50459-8-O |
Leishmania Donovani (cluster #8 Of 8), Other |
Other |
2680 |
0.43 |
Functional ≤ 10μM
|
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
220 |
0.52 |
Functional ≤ 10μM
|
|
Z50725-7-O |
Trypanosoma Brucei Rhodesiense (cluster #7 Of 7), Other |
Other |
600 |
0.48 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
1500 |
0.45 |
Functional ≤ 10μM
|
|
Z80695-2-O |
BE (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
1280 |
0.46 |
Functional ≤ 10μM
|
|
Z81240-1-O |
MAC15A (Colon Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
3950 |
0.42 |
Functional ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
1050 |
0.47 |
ADME/T ≤ 10μM |
|
Z81057-4-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #4 Of 5), Other |
Other |
1180 |
0.46 |
ADME/T ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
1120 |
0.46 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
0.21 |
-16.41 |
0 |
2 |
0 |
17 |
232.286 |
0 |
↓
|
|