|
|
Analogs
-
3984042
-
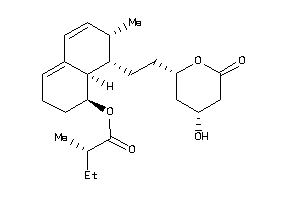
-
4245612
-
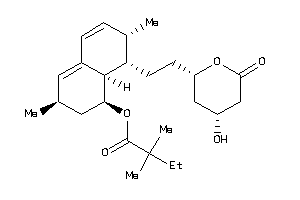
-
4245664
-
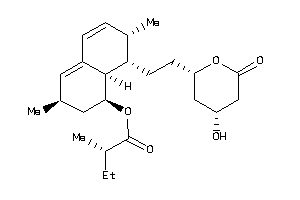
-
4521332
-
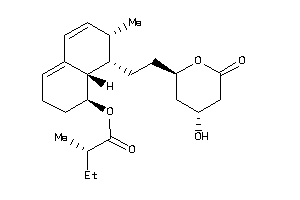
-
26504552
-
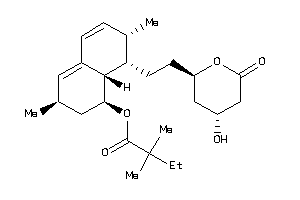
Draw
Identity
99%
90%
80%
70%
Vendors
And 51 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.39 |
Binding ≤ 10μM
|
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.39 |
Binding ≤ 10μM
|
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
27 |
0.37 |
Binding ≤ 10μM |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Functional ≤ 10μM
|
|
ICAM1-2-E |
Intercellular Adhesion Molecule-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3780 |
0.26 |
Functional ≤ 10μM
|
|
ITAL-2-E |
Leukocyte Adhesion Glycoprotein LFA-1 Alpha (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3780 |
0.26 |
Functional ≤ 10μM
|
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
25 |
0.37 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
7100 |
0.25 |
Functional ≤ 10μM
|
|
Z80952-1-O |
HES (cluster #1 Of 1), Other |
Other |
17 |
0.38 |
Functional ≤ 10μM
|
|
Z81020-2-O |
HepG2 (Hepatoblastoma Cells) (cluster #2 Of 8), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z81020-2-O |
HepG2 (Hepatoblastoma Cells) (cluster #2 Of 8), Other |
Other |
79 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
11.28 |
-12.84 |
1 |
5 |
0 |
73 |
404.547 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
7100 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.01 |
0.86 |
-6.83 |
4 |
6 |
0 |
107 |
368.47 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
7100 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.01 |
1.34 |
-7.57 |
4 |
6 |
0 |
107 |
368.47 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
10.68 |
-12.93 |
1 |
5 |
0 |
73 |
390.52 |
7 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
4.5 |
-7.27 |
0 |
10 |
0 |
129 |
236.136 |
4 |
↓
|
|
|
|
Analogs
-
2581981
-
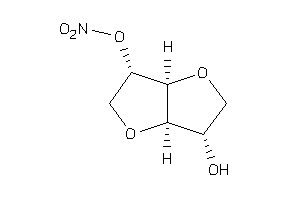
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
4.5 |
-7.29 |
0 |
10 |
0 |
129 |
236.136 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
3.85 |
-4.55 |
0 |
10 |
0 |
129 |
236.136 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.38 |
Binding ≤ 10μM
|
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
5 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
11.12 |
-14.04 |
1 |
5 |
0 |
73 |
404.547 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM4-2-E |
Muscarinic Acetylcholine Receptor M4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4466 |
0.62 |
Functional ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Functional ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
8260 |
0.59 |
Binding ≤ 10μM
|
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
280 |
0.76 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.04 |
5.73 |
-35.78 |
1 |
3 |
1 |
31 |
170.232 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM4-2-E |
Muscarinic Acetylcholine Receptor M4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4466 |
0.62 |
Functional ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Functional ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
8260 |
0.59 |
Binding ≤ 10μM
|
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
280 |
0.76 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.04 |
5.73 |
-35.74 |
1 |
3 |
1 |
31 |
170.232 |
2 |
↓
|
|
|
|
Analogs
-
2575028
-
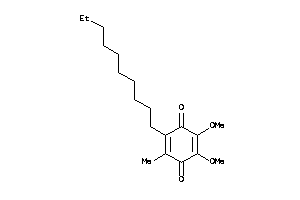
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
1300 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.20 |
1.23 |
-8.88 |
1 |
5 |
0 |
72 |
338.444 |
12 |
↓
|
|
|
|
Analogs
-
3881479
-
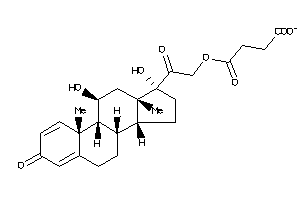
-
3881478
-
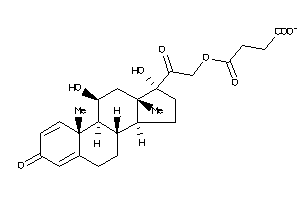
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
-0.24 |
-53.02 |
2 |
8 |
-1 |
141 |
473.542 |
7 |
↓
|
|
|
|
Analogs
-
3881479
-

-
3881478
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
8.95 |
-122.64 |
2 |
8 |
0 |
141 |
473.542 |
7 |
↓
|
|
|
|
Analogs
-
3881479
-

-
3881478
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
1.3 |
-54.56 |
2 |
8 |
-1 |
141 |
473.542 |
7 |
↓
|
|
|
|
Analogs
-
3881479
-

-
3881478
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
0.01 |
-53.43 |
2 |
8 |
-1 |
141 |
473.542 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
-1.98 |
-51.87 |
2 |
5 |
-1 |
89 |
373.469 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
7.35 |
-54.7 |
2 |
5 |
-1 |
90 |
373.469 |
6 |
↓
|
|
|
|
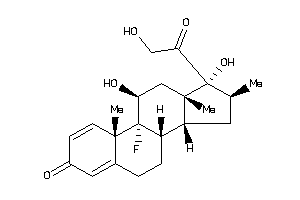
Analogs
-
3978012
-
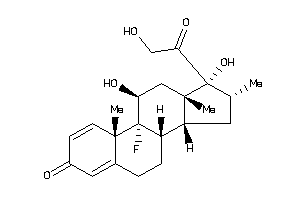
-
3978013
-
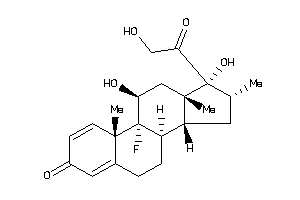
-
3978014
-
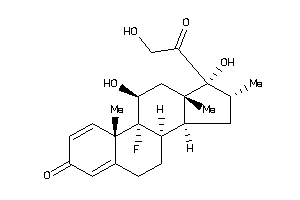
-
3978043
-
-
3978044
-
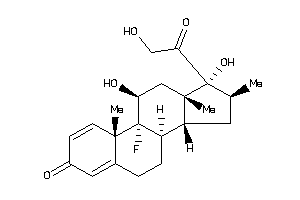
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
22 |
0.38 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Functional ≤ 10μM
|
|
Z50594-7-O |
Mus Musculus (cluster #7 Of 9), Other |
Other |
5 |
0.42 |
Functional ≤ 10μM
|
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
20 |
0.38 |
Functional ≤ 10μM
|
|
Z80178-2-O |
J774.A1 (Macrophage Cells) (cluster #2 Of 3), Other |
Other |
48 |
0.37 |
Functional ≤ 10μM
|
|
Z80301-2-O |
N9 (cluster #2 Of 2), Other |
Other |
74 |
0.36 |
Functional ≤ 10μM
|
|
Z80418-3-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #3 Of 9), Other |
Other |
860 |
0.30 |
Functional ≤ 10μM
|
|
Z80548-4-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #4 Of 5), Other |
Other |
7 |
0.41 |
Functional ≤ 10μM
|
|
Z80954-3-O |
HFF (Foreskin Fibroblasts) (cluster #3 Of 4), Other |
Other |
2 |
0.43 |
Functional ≤ 10μM
|
|
Z81247-4-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #4 Of 9), Other |
Other |
17 |
0.39 |
Functional ≤ 10μM
|
|
Z81338-2-O |
T-cells (cluster #2 Of 3), Other |
Other |
5 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
3.98 |
-16.38 |
3 |
5 |
0 |
95 |
392.467 |
2 |
↓
|
|