|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.42 |
2.83 |
-58.8 |
2 |
3 |
0 |
57 |
127.143 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-4-O |
Rattus Norvegicus (cluster #4 Of 5), Other |
Other |
1300 |
0.75 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.48 |
2.9 |
-61.59 |
2 |
5 |
-1 |
97 |
158.133 |
2 |
↓
|
|
|
|
Analogs
-
5855933
-
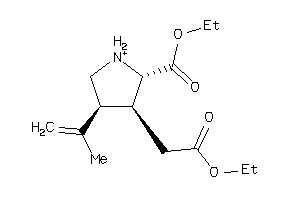
-
34599078
-
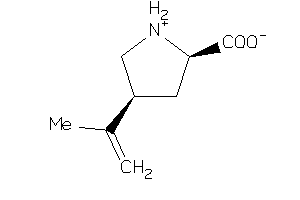
-
1481864
-
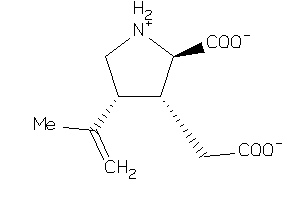
-
2008786
-
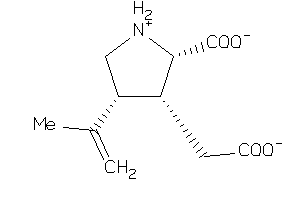
-
3872987
-
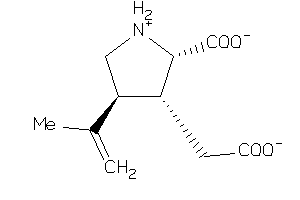
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-6-E |
Adenosine Receptor A3 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
77 |
0.66 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7500 |
0.48 |
Binding ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.49 |
Binding ≤ 10μM
|
|
GRIA3-3-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.49 |
Binding ≤ 10μM
|
|
GRIA4-3-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1714 |
0.54 |
Binding ≤ 10μM
|
|
GRIK1-3-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
177 |
0.63 |
Binding ≤ 10μM
|
|
GRIK2-2-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
53 |
0.68 |
Binding ≤ 10μM
|
|
GRIK3-2-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.75 |
Binding ≤ 10μM
|
|
GRIK4-2-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.76 |
Binding ≤ 10μM
|
|
GRIK5-2-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.76 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7510 |
0.48 |
Functional ≤ 10μM
|
|
GRIK2-2-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.57 |
Functional ≤ 10μM
|
|
Z50597-4-O |
Rattus Norvegicus (cluster #4 Of 5), Other |
Other |
18 |
0.72 |
Binding ≤ 10μM
|
|
Z50466-5-O |
Trypanosoma Cruzi (cluster #5 Of 8), Other |
Other |
3000 |
0.52 |
Functional ≤ 10μM
|
|
Z50597-6-O |
Rattus Norvegicus (cluster #6 Of 12), Other |
Other |
3700 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.81 |
5.21 |
-88.22 |
2 |
5 |
-1 |
97 |
212.225 |
4 |
↓
|
|
|
|
Analogs
-
3861497
-
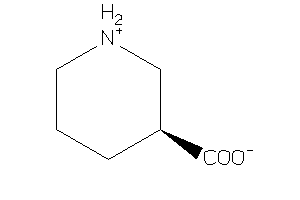
Draw
Identity
99%
90%
80%
70%
Vendors
And 108 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-6-E |
Adenosine Receptor A3 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
10 |
1.24 |
Binding ≤ 10μM
|
|
SC6A1-2-E |
GABA Transporter 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.79 |
Binding ≤ 10μM
|
|
SC6A1-2-E |
GABA Transporter 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8511 |
0.79 |
Binding ≤ 10μM
|
|
S6A11-2-E |
GABA Transporter 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.81 |
Functional ≤ 10μM
|
|
S6A12-1-E |
Betaine Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.81 |
Functional ≤ 10μM
|
|
S6A13-2-E |
GABA Transporter 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.81 |
Functional ≤ 10μM
|
|
SC6A1-1-E |
GABA Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.81 |
Functional ≤ 10μM
|
|
Z50597-4-O |
Rattus Norvegicus (cluster #4 Of 5), Other |
Other |
5200 |
0.82 |
Binding ≤ 10μM
|
|
Z50597-7-O |
Rattus Norvegicus (cluster #7 Of 12), Other |
Other |
5000 |
0.82 |
Functional ≤ 10μM
|
|
Z50597-7-O |
Rattus Norvegicus (cluster #7 Of 12), Other |
Other |
8000 |
0.79 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.23 |
2.96 |
-53.77 |
2 |
3 |
0 |
57 |
129.159 |
1 |
↓
|
|
|
|
Analogs
-
2141051
-
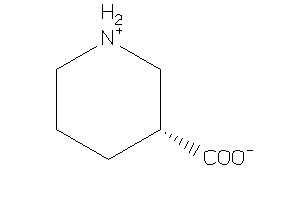
Draw
Identity
99%
90%
80%
70%
Vendors
And 102 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-6-E |
Adenosine Receptor A3 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
10 |
1.24 |
Binding ≤ 10μM
|
|
SC6A1-2-E |
GABA Transporter 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4960 |
0.83 |
Binding ≤ 10μM
|
|
SC6A1-2-E |
GABA Transporter 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.79 |
Binding ≤ 10μM
|
|
S6A11-2-E |
GABA Transporter 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.81 |
Functional ≤ 10μM
|
|
S6A12-1-E |
Betaine Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.81 |
Functional ≤ 10μM
|
|
S6A13-2-E |
GABA Transporter 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.81 |
Functional ≤ 10μM
|
|
SC6A1-1-E |
GABA Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.81 |
Functional ≤ 10μM
|
|
Z50597-4-O |
Rattus Norvegicus (cluster #4 Of 5), Other |
Other |
5200 |
0.82 |
Binding ≤ 10μM
|
|
Z50597-7-O |
Rattus Norvegicus (cluster #7 Of 12), Other |
Other |
8000 |
0.79 |
Functional ≤ 10μM
|
|
Z50597-7-O |
Rattus Norvegicus (cluster #7 Of 12), Other |
Other |
9880 |
0.78 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.23 |
3 |
-54.83 |
2 |
3 |
0 |
57 |
129.159 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.13 |
14.8 |
-62.14 |
1 |
4 |
0 |
54 |
473.413 |
9 |
↓
|
|