|
|
Analogs
-
3978047
-
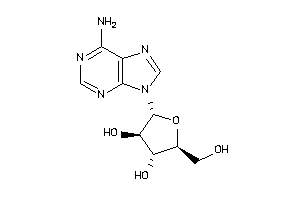
-
3978049
-
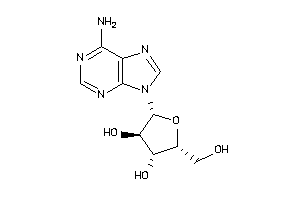
-
4048240
-
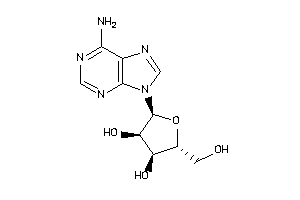
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP4-1-E |
Dipeptidyl Peptidase IV (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
62 |
0.53 |
Binding ≤ 10μM
|
|
Z102131-1-O |
Vaccinia Virus Copenhagen (cluster #1 Of 1), Other |
Other |
2000 |
0.42 |
Functional ≤ 10μM |
|
Z50599-1-O |
Cowpox Virus (cluster #1 Of 1), Other |
Other |
1400 |
0.43 |
Functional ≤ 10μM
|
|
Z50600-1-O |
Vaccinia Virus (cluster #1 Of 2), Other |
Other |
8300 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.85 |
-5.28 |
-13.98 |
5 |
9 |
0 |
140 |
267.245 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z101697-1-O |
Camelpox Virus (cluster #1 Of 1), Other |
Other |
8800 |
0.39 |
Functional ≤ 10μM
|
|
Z101794-1-O |
Felid Herpesvirus 1 (cluster #1 Of 1), Other |
Other |
1300 |
0.46 |
Functional ≤ 10μM |
|
Z101848-1-O |
Human Herpesvirus 8 (cluster #1 Of 1), Other |
Other |
64 |
0.56 |
Functional ≤ 10μM
|
|
Z102080-1-O |
Simplexvirus (cluster #1 Of 1), Other |
Other |
3300 |
0.43 |
Functional ≤ 10μM
|
|
Z102131-1-O |
Vaccinia Virus Copenhagen (cluster #1 Of 1), Other |
Other |
9800 |
0.39 |
Functional ≤ 10μM
|
|
Z50515-2-O |
Human Herpesvirus 2 (cluster #2 Of 2), Other |
Other |
9500 |
0.39 |
Functional ≤ 10μM
|
|
Z50517-1-O |
Human Herpesvirus 5 Strain AD169 (cluster #1 Of 2), Other |
Other |
95 |
0.55 |
Functional ≤ 10μM
|
|
Z50518-3-O |
Human Herpesvirus 4 (cluster #3 Of 5), Other |
Other |
480 |
0.49 |
Functional ≤ 10μM
|
|
Z50527-3-O |
Human Herpesvirus 3 (cluster #3 Of 3), Other |
Other |
70 |
0.56 |
Functional ≤ 10μM |
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
940 |
0.47 |
Functional ≤ 10μM |
|
Z50599-1-O |
Cowpox Virus (cluster #1 Of 1), Other |
Other |
9000 |
0.39 |
Functional ≤ 10μM
|
|
Z50600-1-O |
Vaccinia Virus (cluster #1 Of 2), Other |
Other |
9400 |
0.39 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
570 |
0.49 |
Functional ≤ 10μM
|
|
Z50604-1-O |
Murid Herpesvirus 1 (cluster #1 Of 1), Other |
Other |
40 |
0.58 |
Functional ≤ 10μM
|
|
Z50624-1-O |
Human Adenovirus Type 2 (cluster #1 Of 1), Other |
Other |
3100 |
0.43 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
10000 |
0.39 |
Functional ≤ 10μM
|
|
Z80335-1-O |
NHDF (cluster #1 Of 2), Other |
Other |
500 |
0.49 |
Functional ≤ 10μM
|
|
Z80709-1-O |
Osteoclast-like (Bone Marrow Cells) (cluster #1 Of 1), Other |
Other |
10000 |
0.39 |
Functional ≤ 10μM
|
|
Z80942-1-O |
HEL (Embryonic Lung Cells) (cluster #1 Of 2), Other |
Other |
600 |
0.48 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 1), Other |
Other |
1900 |
0.44 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.70 |
-5.05 |
-52.83 |
4 |
9 |
-1 |
151 |
278.181 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.35 |
-6.46 |
-25.45 |
5 |
7 |
0 |
122 |
259.287 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.83 |
-2.49 |
-17.31 |
3 |
9 |
0 |
152 |
304.262 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.37 |
-5.15 |
-48.8 |
2 |
9 |
-1 |
155 |
303.254 |
3 |
↓
|
|
|
|
Analogs
-
6091017
-
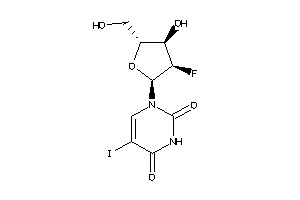
-
6524085
-
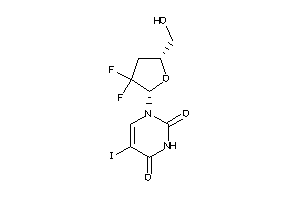
-
22059318
-
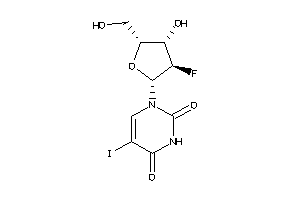
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KITH-1-E |
Thymidine Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
950 |
0.47 |
Binding ≤ 10μM
|
|
Z50599-1-O |
Cowpox Virus (cluster #1 Of 1), Other |
Other |
240 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.79 |
-8.38 |
-11.74 |
3 |
7 |
0 |
104 |
372.09 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z101697-1-O |
Camelpox Virus (cluster #1 Of 1), Other |
Other |
500 |
0.44 |
Functional ≤ 10μM
|
|
Z102131-1-O |
Vaccinia Virus Copenhagen (cluster #1 Of 1), Other |
Other |
2700 |
0.39 |
Functional ≤ 10μM
|
|
Z102132-1-O |
Vaccinia Virus WR (cluster #1 Of 1), Other |
Other |
6500 |
0.36 |
Functional ≤ 10μM
|
|
Z50517-1-O |
Human Herpesvirus 5 Strain AD169 (cluster #1 Of 2), Other |
Other |
80 |
0.50 |
Functional ≤ 10μM
|
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
820 |
0.43 |
Functional ≤ 10μM
|
|
Z50599-1-O |
Cowpox Virus (cluster #1 Of 1), Other |
Other |
4000 |
0.38 |
Functional ≤ 10μM
|
|
Z50599-1-O |
Cowpox Virus (cluster #1 Of 1), Other |
Other |
5000 |
0.37 |
Functional ≤ 10μM
|
|
Z50600-1-O |
Vaccinia Virus (cluster #1 Of 2), Other |
Other |
2700 |
0.39 |
Functional ≤ 10μM
|
|
Z50600-1-O |
Vaccinia Virus (cluster #1 Of 2), Other |
Other |
3500 |
0.38 |
Functional ≤ 10μM
|
|
Z50604-1-O |
Murid Herpesvirus 1 (cluster #1 Of 1), Other |
Other |
160 |
0.48 |
Functional ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
1200 |
0.41 |
Functional ≤ 10μM
|
|
Z50624-1-O |
Human Adenovirus Type 2 (cluster #1 Of 1), Other |
Other |
630 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.63 |
-1.53 |
-55.96 |
4 |
10 |
-1 |
159 |
302.207 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.63 |
-0.38 |
-140.45 |
3 |
10 |
-2 |
162 |
301.199 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.37 |
-1.31 |
-16.11 |
3 |
10 |
0 |
155 |
351.315 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.92 |
-3.79 |
-50.61 |
2 |
10 |
-1 |
158 |
350.307 |
6 |
↓
|
|
|
|
Analogs
-
537850
-
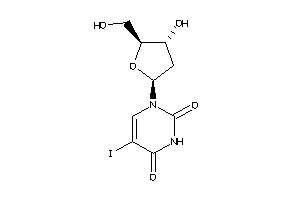
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.80 |
-3.35 |
-14.11 |
3 |
7 |
0 |
105 |
354.1 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.34 |
-6.06 |
-51.55 |
2 |
7 |
-1 |
108 |
353.092 |
2 |
↓
|
|
|
|
Analogs
-
2014826
-
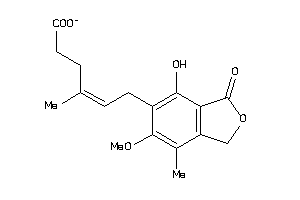
Draw
Identity
99%
90%
80%
70%
Vendors
And 44 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
IMDH1-1-E |
Inosine-5'-monophosphate Dehydrogenase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
55 |
0.44 |
Binding ≤ 10μM
|
|
IMDH2-1-E |
Inosine-5'-monophosphate Dehydrogenase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.50 |
Binding ≤ 10μM
|
|
Z101767-1-O |
Dengue Virus 2 (cluster #1 Of 1), Other |
Other |
5700 |
0.32 |
Functional ≤ 10μM
|
|
Z50442-2-O |
Candida Albicans (cluster #2 Of 4), Other |
Other |
8000 |
0.31 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
58 |
0.44 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
1000 |
0.37 |
Functional ≤ 10μM
|
|
Z50599-1-O |
Cowpox Virus (cluster #1 Of 1), Other |
Other |
3200 |
0.33 |
Functional ≤ 10μM
|
|
Z50641-2-O |
West Nile Virus (cluster #2 Of 2), Other |
Other |
80 |
0.43 |
Functional ≤ 10μM
|
|
Z50678-1-O |
Respiratory Syncytial Virus (cluster #1 Of 2), Other |
Other |
600 |
0.38 |
Functional ≤ 10μM
|
|
Z80040-1-O |
BHK-21 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
40 |
0.45 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
900 |
0.37 |
Functional ≤ 10μM |
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
80 |
0.43 |
Functional ≤ 10μM
|
|
Z80193-1-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #1 Of 12), Other |
Other |
200 |
0.41 |
Functional ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
500 |
0.38 |
Functional ≤ 10μM |
|
Z80401-1-O |
PSN1 (cluster #1 Of 2), Other |
Other |
700 |
0.37 |
Functional ≤ 10μM |
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
80 |
0.43 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
400 |
0.39 |
Functional ≤ 10μM |
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
900 |
0.37 |
Functional ≤ 10μM |
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
99 |
0.43 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
128 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
7.03 |
-53.53 |
1 |
6 |
-1 |
96 |
319.333 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.63 |
7.8 |
-109.76 |
0 |
6 |
-2 |
99 |
318.325 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.63 |
5.05 |
-14.55 |
2 |
6 |
0 |
93 |
320.341 |
6 |
↓
|
|
|
|
|