|
|
|
|
|
Analogs
-
8762249
-
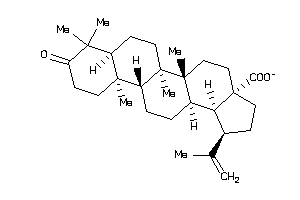
-
8762252
-
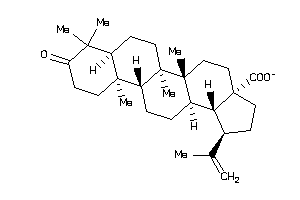
-
8835967
-
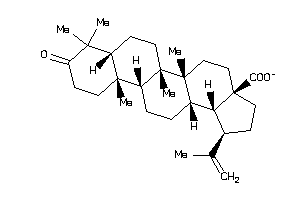
-
8835968
-
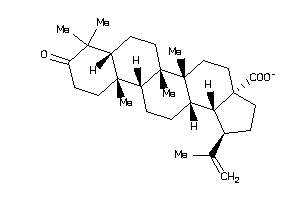
-
8952537
-
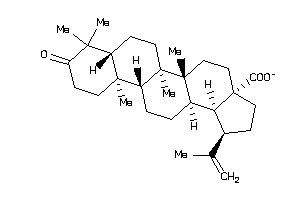
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4710 |
0.23 |
Functional ≤ 10μM
|
|
NR1H4-2-E |
Bile Acid Receptor FXR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z100499-2-O |
SARS Coronavirus (cluster #2 Of 2), Other |
Other |
630 |
0.26 |
Functional ≤ 10μM
|
|
Z103205-2-O |
A431 (cluster #2 Of 4), Other |
Other |
3260 |
0.23 |
Functional ≤ 10μM
|
|
Z50602-3-O |
Human Herpesvirus 1 (cluster #3 Of 5), Other |
Other |
2500 |
0.24 |
Functional ≤ 10μM
|
|
Z50652-2-O |
Influenza A Virus (cluster #2 Of 4), Other |
Other |
5700 |
0.22 |
Functional ≤ 10μM
|
|
Z80186-4-O |
K562 (Erythroleukemia Cells) (cluster #4 Of 11), Other |
Other |
6000 |
0.22 |
Functional ≤ 10μM
|
|
Z80928-4-O |
HCT-116 (Colon Carcinoma Cells) (cluster #4 Of 9), Other |
Other |
2610 |
0.24 |
Functional ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
2980 |
0.23 |
Functional ≤ 10μM
|
|
Z81252-4-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #4 Of 11), Other |
Other |
9770 |
0.21 |
Functional ≤ 10μM
|
|
Z80186-2-O |
K562 (Erythroleukemia Cells) (cluster #2 Of 3), Other |
Other |
6000 |
0.22 |
ADME/T ≤ 10μM
|
|
Z81115-3-O |
KB (Squamous Cell Carcinoma) (cluster #3 Of 3), Other |
Other |
3800 |
0.23 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.86 |
14.43 |
-48.14 |
0 |
3 |
-1 |
57 |
453.687 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.86 |
12.66 |
-7.5 |
1 |
3 |
0 |
54 |
454.695 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
3206938
-
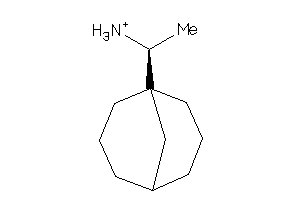
-
3206939
-
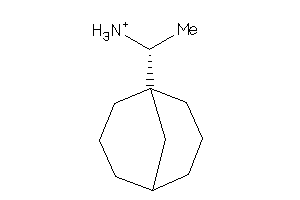
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
-1.33 |
-39.51 |
3 |
1 |
1 |
27 |
180.315 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-3-O |
Trypanosoma Brucei (cluster #3 Of 6), Other |
Other |
5280 |
0.49 |
Functional ≤ 10μM |
|
Z50652-2-O |
Influenza A Virus (cluster #2 Of 4), Other |
Other |
160 |
0.63 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
6.43 |
-42.28 |
2 |
1 |
1 |
17 |
206.353 |
0 |
↓
|
|
|
|
Analogs
-
4159387
-
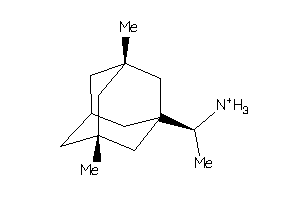
-
4159388
-
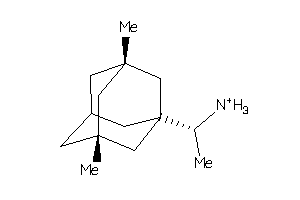
-
40865734
-
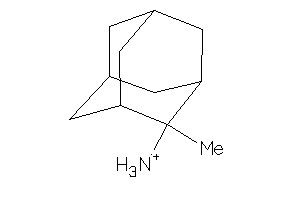
Draw
Identity
99%
90%
80%
70%
Vendors
And 87 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
5.01 |
-42.88 |
3 |
1 |
1 |
28 |
180.315 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
|