|
|
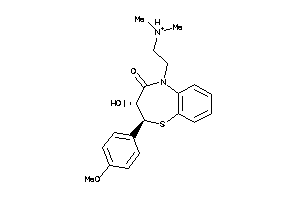
Analogs
-
22056034
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50740-1-O |
Cricetinae Gen. Sp. (cluster #1 Of 2), Other |
Other |
2460 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
-0.17 |
-45.27 |
2 |
5 |
1 |
54 |
373.498 |
5 |
↓
|
|
|
|
Analogs
-
22056034
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50740-1-O |
Cricetinae Gen. Sp. (cluster #1 Of 2), Other |
Other |
2460 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
-0.47 |
-40.08 |
2 |
5 |
1 |
54 |
373.498 |
5 |
↓
|
|
|
|
|
|
|
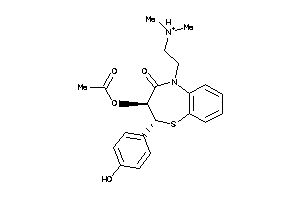
Analogs
-
22056232
-
-
601259
-
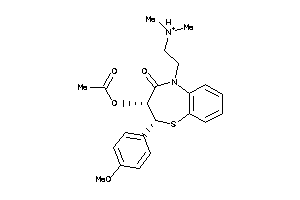
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50740-1-O |
Cricetinae Gen. Sp. (cluster #1 Of 2), Other |
Other |
3270 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
0.8 |
-44.97 |
2 |
6 |
1 |
72 |
401.508 |
7 |
↓
|
|
|
|
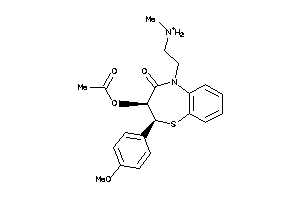
Analogs
-
5166438
-
-
22056232
-

-
601259
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 45 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
54 |
0.35 |
Binding ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4700 |
0.26 |
Binding ≤ 10μM
|
|
CAC1E-1-E |
Voltage-gated R-type Calcium Channel Alpha-1E Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.26 |
Binding ≤ 10μM
|
|
CAC1F-1-E |
Voltage-gated L-type Calcium Channel Alpha-1F Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
380 |
0.31 |
Binding ≤ 10μM |
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
380 |
0.31 |
Binding ≤ 10μM |
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4760 |
0.26 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
59 |
0.35 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
59 |
0.35 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.30 |
ADME/T ≤ 10μM
|
|
Z50592-1-O |
Oryctolagus Cuniculus (cluster #1 Of 1), Other |
Other |
1000 |
0.29 |
Binding ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
95 |
0.34 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
210 |
0.32 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
790 |
0.29 |
Functional ≤ 10μM
|
|
Z50740-1-O |
Cricetinae Gen. Sp. (cluster #1 Of 2), Other |
Other |
980 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
12.1 |
-42.54 |
1 |
6 |
1 |
60 |
415.535 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.34 |
9.64 |
-12.57 |
0 |
6 |
0 |
59 |
414.527 |
7 |
↓
|
|
|
|
Analogs
-
1851916
-
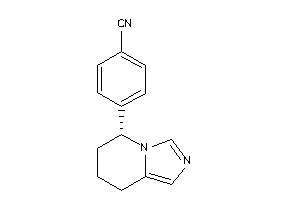
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
2.5 |
-9.93 |
0 |
3 |
0 |
41 |
223.279 |
1 |
↓
|
|
|
|
Analogs
-
6398
-
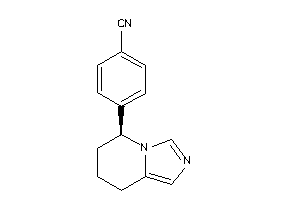
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C11B1-1-E |
Cytochrome P450 11B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.66 |
Binding ≤ 10μM
|
|
C11B1-1-E |
Cytochrome P450 11B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.61 |
Binding ≤ 10μM
|
|
C11B2-2-E |
Cytochrome P450 11B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.74 |
Binding ≤ 10μM
|
|
C11B2-2-E |
Cytochrome P450 11B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
171 |
0.56 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
52 |
0.60 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
680 |
0.51 |
Binding ≤ 10μM
|
|
C11B1-1-E |
Cytochrome P450 11B1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.49 |
Functional ≤ 10μM
|
|
C11B2-1-E |
Cytochrome P450 11B2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.51 |
Functional ≤ 10μM
|
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.62 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
600 |
0.51 |
Functional ≤ 10μM
|
|
Z50740-1-O |
Cricetinae Gen. Sp. (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
9.77 |
-36.47 |
1 |
3 |
1 |
43 |
224.287 |
1 |
↓
|
|