|
|
Analogs
-
33826966
-
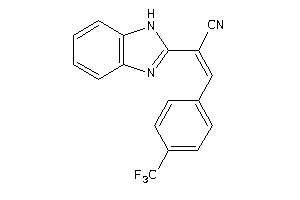
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
9110 |
0.31 |
Functional ≤ 10μM
|
|
Z81146-1-O |
LCLC-103H Cell Line (cluster #1 Of 1), Other |
Other |
1600 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.28 |
2.99 |
-8.73 |
1 |
3 |
0 |
52 |
313.282 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.40 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z100501-1-O |
Lu1 (Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
80 |
0.52 |
Functional ≤ 10μM
|
|
Z103204-1-O |
A427 (cluster #1 Of 4), Other |
Other |
890 |
0.45 |
Functional ≤ 10μM
|
|
Z103205-3-O |
A431 (cluster #3 Of 4), Other |
Other |
470 |
0.47 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
501 |
0.46 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
73 |
0.53 |
Functional ≤ 10μM
|
|
Z80008-2-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
890 |
0.45 |
Functional ≤ 10μM
|
|
Z80036-2-O |
BaF3 (IL-3-dependent Pro-B-cells) (cluster #2 Of 3), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z80145-1-O |
H69 (cluster #1 Of 4), Other |
Other |
1800 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-10-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #10 Of 12), Other |
Other |
660 |
0.46 |
Functional ≤ 10μM
|
|
Z80164-3-O |
HT-1080 (Fibrosarcoma Cells) (cluster #3 Of 6), Other |
Other |
4300 |
0.40 |
Functional ≤ 10μM
|
|
Z80186-10-O |
K562 (Erythroleukemia Cells) (cluster #10 Of 11), Other |
Other |
380 |
0.47 |
Functional ≤ 10μM
|
|
Z80193-4-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #4 Of 12), Other |
Other |
500 |
0.46 |
Functional ≤ 10μM |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
430 |
0.47 |
Functional ≤ 10μM
|
|
Z80322-1-O |
NCI-H358 (Lung Carcinama Cells) (cluster #1 Of 2), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z80509-1-O |
SNU-638 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.42 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
800 |
0.45 |
Functional ≤ 10μM
|
|
Z80682-7-O |
A549 (Lung Carcinoma Cells) (cluster #7 Of 11), Other |
Other |
470 |
0.47 |
Functional ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2300 |
0.42 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
410 |
0.47 |
Functional ≤ 10μM
|
|
Z81024-8-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #8 Of 8), Other |
Other |
510 |
0.46 |
Functional ≤ 10μM
|
|
Z81115-3-O |
KB (Squamous Cell Carcinoma) (cluster #3 Of 6), Other |
Other |
812 |
0.45 |
Functional ≤ 10μM
|
|
Z81115-3-O |
KB (Squamous Cell Carcinoma) (cluster #3 Of 6), Other |
Other |
1780 |
0.42 |
Functional ≤ 10μM |
|
Z81170-5-O |
LNCaP (Prostate Carcinoma) (cluster #5 Of 5), Other |
Other |
3250 |
0.40 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
310 |
0.48 |
Functional ≤ 10μM
|
|
Z81252-10-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #10 Of 11), Other |
Other |
2300 |
0.42 |
Functional ≤ 10μM
|
|
Z81277-1-O |
NCI-N417 (cluster #1 Of 2), Other |
Other |
870 |
0.45 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 4), Other |
Other |
2600 |
0.41 |
ADME/T ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 1), Other |
Other |
5010 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
190 |
0.50 |
ADME/T ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
3820 |
0.40 |
ADME/T ≤ 10μM |
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
5500 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
2070 |
0.42 |
ADME/T ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1820 |
0.42 |
ADME/T ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 1), Other |
Other |
3 |
0.63 |
ADME/T ≤ 10μM
|
|
Z81057-4-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #4 Of 5), Other |
Other |
370 |
0.47 |
ADME/T ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
5300 |
0.39 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.28 |
8.05 |
-7.84 |
1 |
2 |
0 |
29 |
246.313 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.28 |
8.52 |
-35.14 |
2 |
2 |
1 |
30 |
247.321 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
|
CLK3-1-E |
Dual Specificity Protein Kinase CLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
41 |
0.41 |
Binding ≤ 10μM
|
|
CSK21-1-E |
Casein Kinase II Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM
|
|
DAPK3-1-E |
Death-associated Protein Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
17 |
0.44 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
55 |
0.41 |
Binding ≤ 10μM
|
|
HIPK3-1-E |
Homeodomain-interacting Protein Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
45 |
0.41 |
Binding ≤ 10μM
|
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
46 |
0.41 |
Binding ≤ 10μM
|
|
TBK1-1-E |
Serine/threonine-protein Kinase TBK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
35 |
0.42 |
Binding ≤ 10μM
|
|
Z103203-2-O |
A375 (cluster #2 Of 3), Other |
Other |
3900 |
0.30 |
Functional ≤ 10μM
|
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
5300 |
0.30 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
8900 |
0.28 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
2100 |
0.32 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
3000 |
0.31 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
2200 |
0.32 |
Functional ≤ 10μM
|
|
Z81072-6-O |
Jurkat (Acute Leukemic T-cells) (cluster #6 Of 10), Other |
Other |
2500 |
0.31 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
4700 |
0.30 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
6400 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.14 |
10.5 |
-55.54 |
1 |
5 |
-1 |
78 |
348.769 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.84 |
5.03 |
-92.09 |
7 |
8 |
2 |
122 |
456.497 |
5 |
↓
|
|
|
|
Analogs
-
3675826
-
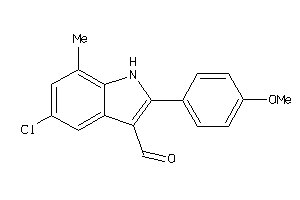
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
0.8 |
-8.21 |
1 |
3 |
0 |
42 |
251.285 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
0.64 |
-10.62 |
0 |
3 |
0 |
43 |
247.253 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKA-3-E |
Serine/threonine-protein Kinase Aurora-A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
31 |
0.44 |
Binding ≤ 10μM |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
1600 |
0.34 |
Functional ≤ 10μM |
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
7800 |
0.30 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
9.25 |
-15.42 |
2 |
6 |
0 |
76 |
317.352 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 75 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYSY-3-E |
Thymidylate Synthase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.80 |
Binding ≤ 10μM
|
|
Z103205-4-O |
A431 (cluster #4 Of 4), Other |
Other |
18 |
1.20 |
Functional ≤ 10μM
|
|
Z103419-1-O |
UACC-812 (cluster #1 Of 2), Other |
Other |
3100 |
0.86 |
Functional ≤ 10μM
|
|
Z50425-14-O |
Plasmodium Falciparum (cluster #14 Of 22), Other |
Other |
7943 |
0.79 |
Functional ≤ 10μM
|
|
Z80035-6-O |
B16 (Melanoma Cells) (cluster #6 Of 7), Other |
Other |
8730 |
0.79 |
Functional ≤ 10μM
|
|
Z80054-2-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #2 Of 4), Other |
Other |
1500 |
0.91 |
Functional ≤ 10μM
|
|
Z80064-6-O |
CCRF-CEM (T-cell Leukemia) (cluster #6 Of 9), Other |
Other |
5000 |
0.82 |
Functional ≤ 10μM
|
|
Z80088-4-O |
CHO (Ovarian Cells) (cluster #4 Of 4), Other |
Other |
5000 |
0.82 |
Functional ≤ 10μM
|
|
Z80101-1-O |
COLO 320DM (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
1100 |
0.93 |
Functional ≤ 10μM |
|
Z80120-3-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
6000 |
0.81 |
Functional ≤ 10μM |
|
Z80125-6-O |
DU-145 (Prostate Carcinoma) (cluster #6 Of 9), Other |
Other |
4800 |
0.83 |
Functional ≤ 10μM
|
|
Z80136-3-O |
FM3A (Breast Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
180 |
1.05 |
Functional ≤ 10μM
|
|
Z80156-6-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #6 Of 12), Other |
Other |
9500 |
0.78 |
Functional ≤ 10μM
|
|
Z80164-4-O |
HT-1080 (Fibrosarcoma Cells) (cluster #4 Of 6), Other |
Other |
9450 |
0.78 |
Functional ≤ 10μM
|
|
Z80166-8-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #8 Of 12), Other |
Other |
7300 |
0.80 |
Functional ≤ 10μM
|
|
Z80186-7-O |
K562 (Erythroleukemia Cells) (cluster #7 Of 11), Other |
Other |
2400 |
0.87 |
Functional ≤ 10μM
|
|
Z80192-2-O |
KM12 (Colon Adenocarcinoma Cells) (cluster #2 Of 4), Other |
Other |
8320 |
0.79 |
Functional ≤ 10μM
|
|
Z80193-8-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #8 Of 12), Other |
Other |
970 |
0.94 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
9600 |
0.78 |
Functional ≤ 10μM
|
|
Z80268-1-O |
MKN-28 (Gastric Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2500 |
0.87 |
Functional ≤ 10μM |
|
Z80269-3-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
3000 |
0.86 |
Functional ≤ 10μM |
|
Z80347-2-O |
NUGC-3 (Gastric Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
4400 |
0.83 |
Functional ≤ 10μM |
|
Z80362-5-O |
P388 (Lymphoma Cells) (cluster #5 Of 8), Other |
Other |
609 |
0.97 |
Functional ≤ 10μM |
|
Z80390-6-O |
PC-3 (Prostate Carcinoma Cells) (cluster #6 Of 10), Other |
Other |
16 |
1.21 |
Functional ≤ 10μM
|
|
Z80485-4-O |
SK-MEL-28 (Melanoma Cells) (cluster #4 Of 6), Other |
Other |
4500 |
0.83 |
Functional ≤ 10μM
|
|
Z80490-4-O |
SK-MES-1 (cluster #4 Of 4), Other |
Other |
310 |
1.01 |
Functional ≤ 10μM
|
|
Z80525-2-O |
SW48 (cluster #2 Of 2), Other |
Other |
5700 |
0.82 |
Functional ≤ 10μM |
|
Z80526-3-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #3 Of 6), Other |
Other |
8100 |
0.79 |
Functional ≤ 10μM
|
|
Z80532-5-O |
T-24 (Bladder Carcinoma Cells) (cluster #5 Of 6), Other |
Other |
4100 |
0.84 |
Functional ≤ 10μM
|
|
Z80583-6-O |
Vero (Kidney Cells) (cluster #6 Of 7), Other |
Other |
6810 |
0.80 |
Functional ≤ 10μM
|
|
Z80641-1-O |
791T Cell Line (cluster #1 Of 1), Other |
Other |
5500 |
0.82 |
Functional ≤ 10μM
|
|
Z80653-2-O |
9L (Glioma Cells) (cluster #2 Of 2), Other |
Other |
1000 |
0.93 |
Functional ≤ 10μM
|
|
Z80682-9-O |
A549 (Lung Carcinoma Cells) (cluster #9 Of 11), Other |
Other |
9200 |
0.78 |
Functional ≤ 10μM
|
|
Z80686-1-O |
AZ-521 Cell Line (cluster #1 Of 1), Other |
Other |
3900 |
0.84 |
Functional ≤ 10μM |
|
Z80712-5-O |
T47D (Breast Carcinoma Cells) (cluster #5 Of 7), Other |
Other |
600 |
0.97 |
Functional ≤ 10μM
|
|
Z80724-1-O |
C170 Cell Line (cluster #1 Of 1), Other |
Other |
2000 |
0.89 |
Functional ≤ 10μM
|
|
Z80742-2-O |
C6 (Glioma Cells) (cluster #2 Of 3), Other |
Other |
6100 |
0.81 |
Functional ≤ 10μM
|
|
Z80874-5-O |
CEM (T-cell Leukemia) (cluster #5 Of 7), Other |
Other |
9230 |
0.78 |
Functional ≤ 10μM
|
|
Z80928-7-O |
HCT-116 (Colon Carcinoma Cells) (cluster #7 Of 9), Other |
Other |
8510 |
0.79 |
Functional ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 8), Other |
Other |
9200 |
0.78 |
Functional ≤ 10μM
|
|
Z81024-6-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #6 Of 8), Other |
Other |
8500 |
0.79 |
Functional ≤ 10μM |
|
Z81034-5-O |
A2780 (Ovarian Carcinoma Cells) (cluster #5 Of 10), Other |
Other |
7210 |
0.80 |
Functional ≤ 10μM
|
|
Z81041-1-O |
Prostatic Carcinoma Cells (cluster #1 Of 2), Other |
Other |
6400 |
0.81 |
Functional ≤ 10μM
|
|
Z81043-2-O |
N592 (cluster #2 Of 2), Other |
Other |
5400 |
0.82 |
Functional ≤ 10μM
|
|
Z81048-1-O |
TSU (Prostatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
3600 |
0.85 |
Functional ≤ 10μM
|
|
Z81115-6-O |
KB (Squamous Cell Carcinoma) (cluster #6 Of 6), Other |
Other |
4460 |
0.83 |
Functional ≤ 10μM
|
|
Z81121-1-O |
KKLS Cell Line (cluster #1 Of 1), Other |
Other |
6200 |
0.81 |
Functional ≤ 10μM |
|
Z81170-4-O |
LNCaP (Prostate Carcinoma) (cluster #4 Of 5), Other |
Other |
4900 |
0.83 |
Functional ≤ 10μM
|
|
Z81247-7-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #7 Of 9), Other |
Other |
9700 |
0.78 |
Functional ≤ 10μM
|
|
Z81252-9-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #9 Of 11), Other |
Other |
9600 |
0.78 |
Functional ≤ 10μM
|
|
Z81330-1-O |
SNU- (cluster #1 Of 1), Other |
Other |
2700 |
0.87 |
Functional ≤ 10μM |
|
Z81331-3-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #3 Of 6), Other |
Other |
9000 |
0.78 |
Functional ≤ 10μM
|
|
Z81335-3-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #3 Of 5), Other |
Other |
9710 |
0.78 |
Functional ≤ 10μM
|
|
Z80156-3-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #3 Of 4), Other |
Other |
160 |
1.06 |
ADME/T ≤ 10μM
|
|
Z80192-2-O |
KM12 (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
7940 |
0.79 |
ADME/T ≤ 10μM
|
|
Z80193-4-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #4 Of 4), Other |
Other |
560 |
0.97 |
ADME/T ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
200 |
1.04 |
ADME/T ≤ 10μM
|
|
Z80901-2-O |
HaCaT (Keratinocytes) (cluster #2 Of 2), Other |
Other |
400 |
1.00 |
ADME/T ≤ 10μM
|
|
Z80928-2-O |
HCT-116 (Colon Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
2040 |
0.89 |
ADME/T ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 3), Other |
Other |
570 |
0.97 |
ADME/T ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
6610 |
0.81 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.59 |
-1.23 |
-13.33 |
2 |
4 |
0 |
66 |
130.078 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.13 |
-4 |
-48.58 |
1 |
4 |
-1 |
69 |
129.07 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.13 |
-3.14 |
-32.57 |
1 |
4 |
-1 |
69 |
129.07 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
190 |
0.45 |
Functional ≤ 10μM
|
|
Z81252-10-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #10 Of 11), Other |
Other |
430 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
3.19 |
-9.65 |
1 |
3 |
0 |
63 |
269.307 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.03 |
1.88 |
-78.27 |
2 |
8 |
2 |
87 |
410.474 |
6 |
↓
|
|
|
|
Analogs
-
6815
-
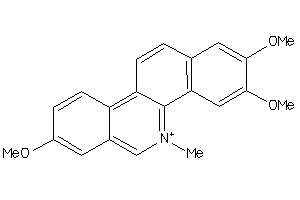
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.00 |
0.77 |
-36.44 |
1 |
5 |
1 |
51 |
350.394 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80012-1-O |
A 172 (Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
|
Z80019-1-O |
A-427 (Lung Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
|
Z80179-1-O |
J82 (Bladder Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
|
Z80532-3-O |
T-24 (Bladder Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
5.62 |
-16.24 |
1 |
3 |
0 |
42 |
251.285 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.51 |
14.97 |
-138.1 |
6 |
9 |
3 |
96 |
596.8 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.51 |
14.61 |
-93.93 |
5 |
9 |
2 |
95 |
595.792 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM
|
|
CHK1-1-E |
Serine/threonine-protein Kinase Chk1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.42 |
Binding ≤ 10μM
|
|
CHK2-1-E |
Serine/threonine-protein Kinase Chk2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.37 |
Binding ≤ 10μM
|
|
E2AK2-1-E |
Interferon-induced, Double-stranded RNA-activated Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM |
|
GSK3B-4-E |
Glycogen Synthase Kinase-3 Beta (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
500 |
0.42 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
10000 |
0.33 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
1000 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.55 |
5.93 |
-10.93 |
2 |
6 |
0 |
83 |
276.255 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.55 |
6.42 |
-44.52 |
3 |
6 |
1 |
84 |
277.263 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
3300 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.19 |
0.49 |
-7.17 |
2 |
2 |
0 |
31 |
258.324 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-2-E |
HERG (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
210 |
0.33 |
Binding ≤ 10μM
|
|
S22A1-1-E |
Solute Carrier Family 22 Member 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9940 |
0.25 |
Binding ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.31 |
Binding ≤ 10μM |
|
TOP1-1-E |
DNA Topoisomerase I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Functional ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Functional ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Functional ≤ 10μM
|
|
ADO-1-E |
Aldehyde Oxidase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6400 |
0.26 |
ADME/T ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.26 |
ADME/T ≤ 10μM
|
|
Z103205-3-O |
A431 (cluster #3 Of 4), Other |
Other |
5890 |
0.26 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
600 |
0.31 |
Functional ≤ 10μM
|
|
Z50460-4-O |
Leishmania Major (cluster #4 Of 4), Other |
Other |
1500 |
0.29 |
Functional ≤ 10μM
|
|
Z80064-8-O |
CCRF-CEM (T-cell Leukemia) (cluster #8 Of 9), Other |
Other |
139 |
0.34 |
Functional ≤ 10μM
|
|
Z80084-1-O |
CEM-VLB (Vinblastine-resistant T-cell Leukemia Cells) (cluster #1 Of 2), Other |
Other |
520 |
0.31 |
Functional ≤ 10μM
|
|
Z80085-1-O |
CEM-VM1 (Etoposide-resistant T-cell Leukemia Cells) (cluster #1 Of 2), Other |
Other |
1910 |
0.29 |
Functional ≤ 10μM
|
|
Z80089-1-O |
CHO-AA8 (cluster #1 Of 2), Other |
Other |
1 |
0.45 |
Functional ≤ 10μM
|
|
Z80115-1-O |
DC3F (cluster #1 Of 1), Other |
Other |
4 |
0.42 |
Functional ≤ 10μM
|
|
Z80117-2-O |
DC3F/AD-II (cluster #2 Of 2), Other |
Other |
47 |
0.37 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
120 |
0.35 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
6 |
0.41 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
559 |
0.31 |
Functional ≤ 10μM |
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
23 |
0.38 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
50 |
0.37 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
75 |
0.36 |
Functional ≤ 10μM
|
|
Z80227-2-O |
MCF7-ADR (Breast Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
1165 |
0.30 |
Functional ≤ 10μM
|
|
Z80244-7-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #7 Of 7), Other |
Other |
490 |
0.32 |
Functional ≤ 10μM
|
|
Z80322-1-O |
NCI-H358 (Lung Carcinama Cells) (cluster #1 Of 2), Other |
Other |
150 |
0.34 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
890 |
0.30 |
Functional ≤ 10μM
|
|
Z80475-3-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #3 Of 3), Other |
Other |
60 |
0.36 |
Functional ≤ 10μM
|
|
Z80601-1-O |
XRS6 (cluster #1 Of 1), Other |
Other |
240 |
0.33 |
Functional ≤ 10μM
|
|
Z80608-4-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #4 Of 4), Other |
Other |
180 |
0.34 |
Functional ≤ 10μM
|
|
Z80612-1-O |
2008 (Ovarian Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
4310 |
0.27 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
3520 |
0.27 |
Functional ≤ 10μM
|
|
Z80712-7-O |
T47D (Breast Carcinoma Cells) (cluster #7 Of 7), Other |
Other |
220 |
0.33 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
1485 |
0.29 |
Functional ≤ 10μM |
|
Z80846-2-O |
F460pv8/eto Cell Line (cluster #2 Of 2), Other |
Other |
40 |
0.37 |
Functional ≤ 10μM
|
|
Z80889-2-O |
NCI-H322M (Non-small Cell Lung Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
210 |
0.33 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
700 |
0.31 |
Functional ≤ 10μM
|
|
Z81024-8-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #8 Of 8), Other |
Other |
60 |
0.36 |
Functional ≤ 10μM
|
|
Z81034-6-O |
A2780 (Ovarian Carcinoma Cells) (cluster #6 Of 10), Other |
Other |
100 |
0.35 |
Functional ≤ 10μM
|
|
Z81072-6-O |
Jurkat (Acute Leukemic T-cells) (cluster #6 Of 10), Other |
Other |
80 |
0.35 |
Functional ≤ 10μM
|
|
Z81160-1-O |
Lewis Lung Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z81164-1-O |
LL Cell Line (cluster #1 Of 2), Other |
Other |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z81167-1-O |
LLTC Cell Line (cluster #1 Of 1), Other |
Other |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
901 |
0.30 |
Functional ≤ 10μM |
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
9500 |
0.25 |
Functional ≤ 10μM
|
|
Z81252-10-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #10 Of 11), Other |
Other |
140 |
0.34 |
Functional ≤ 10μM
|
|
Z81280-1-O |
NCI-H226 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
1010 |
0.30 |
Functional ≤ 10μM
|
|
Z81284-2-O |
NCI-H647 (cluster #2 Of 2), Other |
Other |
70 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.91 |
6.33 |
-44.52 |
2 |
6 |
0 |
84 |
393.468 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.91 |
5.83 |
-16.62 |
2 |
6 |
0 |
80 |
393.468 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.91 |
5.94 |
-47.33 |
1 |
6 |
-1 |
82 |
392.46 |
5 |
↓
|
|
|
|
Analogs
-
6222821
-
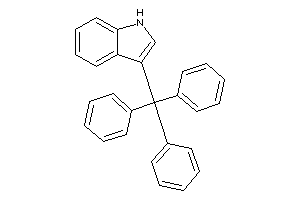
Draw
Identity
99%
90%
80%
70%
Vendors
And 33 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50459-8-O |
Leishmania Donovani (cluster #8 Of 8), Other |
Other |
1250 |
0.43 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
5500 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.20 |
9.06 |
-7.82 |
2 |
2 |
0 |
32 |
246.313 |
2 |
↓
|
|
|
|
|
|
|
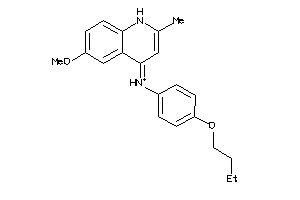
Analogs
-
5808067
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
3 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
-1.95 |
-8.1 |
2 |
4 |
0 |
54 |
294.354 |
4 |
↓
|
|
|
|
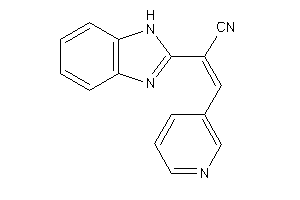
Analogs
-
3899324
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
6.62 |
-8.48 |
1 |
4 |
0 |
65 |
246.273 |
2 |
↓
|
|
|
|
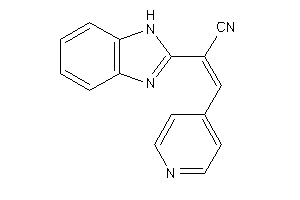
Analogs
-
5387659
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
9000 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
6.61 |
-9.22 |
1 |
4 |
0 |
65 |
246.273 |
2 |
↓
|
|
|
|
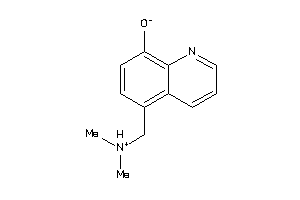
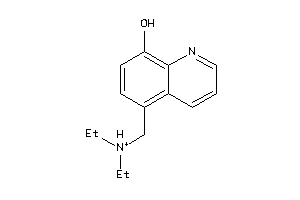
Analogs
-
33724421
-
-
4206713
-
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.52 |
8.63 |
-57.9 |
3 |
5 |
1 |
71 |
422.508 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.52 |
8.42 |
-112.24 |
0 |
5 |
-2 |
75 |
419.484 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.52 |
7.66 |
-61.18 |
1 |
5 |
-1 |
72 |
420.492 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
10.69 |
-11.53 |
1 |
5 |
0 |
64 |
389.842 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NFKB1-1-E |
Nuclear Factor NF-kappa-B P105 Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM
|
|
Z80156-12-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #12 Of 12), Other |
Other |
900 |
0.60 |
Functional ≤ 10μM
|
|
Z80164-6-O |
HT-1080 (Fibrosarcoma Cells) (cluster #6 Of 6), Other |
Other |
2600 |
0.56 |
Functional ≤ 10μM
|
|
Z80186-5-O |
K562 (Erythroleukemia Cells) (cluster #5 Of 11), Other |
Other |
1200 |
0.59 |
Functional ≤ 10μM |
|
Z80211-5-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #5 Of 5), Other |
Other |
900 |
0.60 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
4400 |
0.54 |
Functional ≤ 10μM |
|
Z81072-10-O |
Jurkat (Acute Leukemic T-cells) (cluster #10 Of 10), Other |
Other |
900 |
0.60 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
2.86 |
-14.35 |
0 |
3 |
0 |
43 |
186.166 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
5.12 |
-9.59 |
2 |
3 |
0 |
49 |
244.681 |
1 |
↓
|
|
|
|
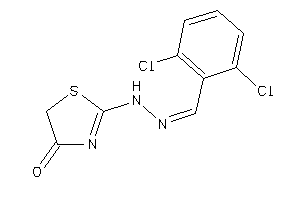
Analogs
-
38194752
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1940 |
0.50 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2750 |
0.49 |
Binding ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
1820 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
5.58 |
-7.97 |
1 |
4 |
0 |
54 |
253.714 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
1.83 |
5.61 |
-8.68 |
1 |
4 |
0 |
54 |
253.714 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
2.36 |
4.64 |
-15.98 |
1 |
4 |
0 |
54 |
253.714 |
3 |
↓
|
|
|
|
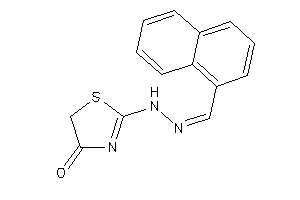
Analogs
-
33909094
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
8190 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.11 |
4.79 |
-15.37 |
1 |
4 |
0 |
54 |
233.296 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.11 |
5.86 |
-31.7 |
2 |
4 |
1 |
55 |
234.304 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
7780 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
4.18 |
-19.24 |
1 |
4 |
0 |
54 |
237.259 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.82 |
5.27 |
-34.97 |
2 |
4 |
1 |
55 |
238.267 |
3 |
↓
|
|
|
|
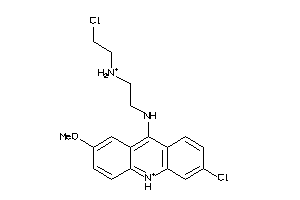
Analogs
-
5733913
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
280 |
0.44 |
Functional ≤ 10μM
|
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
7000 |
0.34 |
Functional ≤ 10μM |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
9300 |
0.34 |
Functional ≤ 10μM |
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
6000 |
0.35 |
Functional ≤ 10μM |
|
Z81252-10-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #10 Of 11), Other |
Other |
8120 |
0.34 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
2070 |
0.38 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
-4.51 |
-93.85 |
5 |
4 |
2 |
63 |
303.793 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-6.37 |
24.54 |
-432.11 |
4 |
8 |
6 |
75 |
680.86 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
71 |
0.45 |
Functional ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
71 |
0.45 |
Functional ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
71 |
0.45 |
Functional ≤ 10μM
|
|
Z80017-2-O |
A2780cisR (Cisplatin-resistant Ovarian Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
570 |
0.40 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
2300 |
0.36 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
990 |
0.38 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
98 |
0.45 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
3800 |
0.34 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
98 |
0.45 |
Functional ≤ 10μM
|
|
Z80493-3-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
980 |
0.38 |
Functional ≤ 10μM
|
|
Z80768-1-O |
CH1 (Ovarian Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
530 |
0.40 |
Functional ≤ 10μM
|
|
Z81034-6-O |
A2780 (Ovarian Carcinoma Cells) (cluster #6 Of 10), Other |
Other |
510 |
0.40 |
Functional ≤ 10μM
|
|
Z81072-6-O |
Jurkat (Acute Leukemic T-cells) (cluster #6 Of 10), Other |
Other |
580 |
0.40 |
Functional ≤ 10μM
|
|
Z81160-1-O |
Lewis Lung Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
190 |
0.43 |
Functional ≤ 10μM
|
|
Z81167-1-O |
LLTC Cell Line (cluster #1 Of 1), Other |
Other |
189 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.34 |
-0.53 |
-51.34 |
2 |
4 |
1 |
46 |
294.378 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 45 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
4.93 |
-9.62 |
3 |
3 |
0 |
55 |
209.252 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
12367925
-
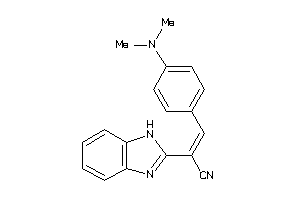
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
4000 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
0.8 |
-9.01 |
1 |
4 |
0 |
55 |
288.354 |
3 |
↓
|
|
|
|
Analogs
-
4066930
-
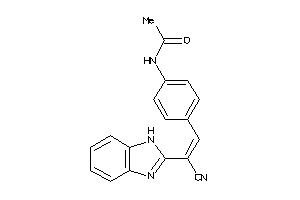
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
4000 |
0.33 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
3000 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
7.31 |
-15.95 |
2 |
5 |
0 |
82 |
302.337 |
3 |
↓
|
|