|
|
Analogs
-
3978074
-
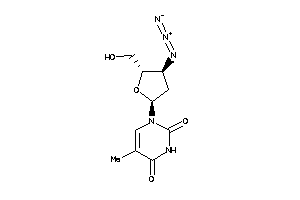
-
4245659
-
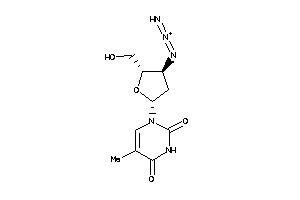
-
4245660
-
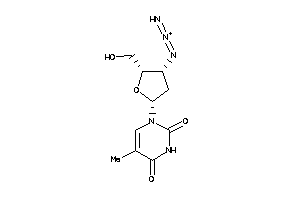
-
4467870
-
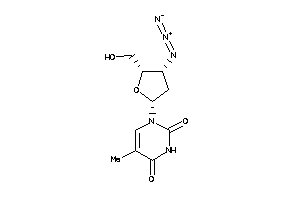
-
11592709
-
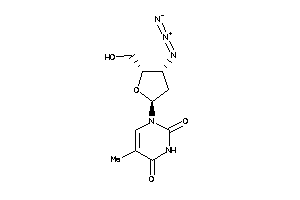
Draw
Identity
99%
90%
80%
70%
Vendors
And 37 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KITH-1-E |
Thymidine Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Binding ≤ 10μM
|
|
CXCR4-1-E |
C-X-C Chemokine Receptor Type 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.60 |
Functional ≤ 10μM
|
|
Z80295-1-O |
MT4 (Lymphocytes) (cluster #1 Of 1), Other |
Other |
20 |
0.57 |
Binding ≤ 10μM
|
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
4 |
0.62 |
Functional ≤ 10μM
|
|
Z102020-1-O |
Porcine Endogenous Retrovirus (cluster #1 Of 1), Other |
Other |
23 |
0.56 |
Functional ≤ 10μM
|
|
Z50459-2-O |
Leishmania Donovani (cluster #2 Of 8), Other |
Other |
3400 |
0.40 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
7 |
0.60 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
46 |
0.54 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
9600 |
0.37 |
Functional ≤ 10μM
|
|
Z50609-1-O |
Simian Immunodeficiency Virus (cluster #1 Of 2), Other |
Other |
12 |
0.58 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
9 |
0.59 |
Functional ≤ 10μM
|
|
Z50664-1-O |
Rauscher Murine Leukemia Virus (cluster #1 Of 1), Other |
Other |
23 |
0.56 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
9 |
0.59 |
Functional ≤ 10μM
|
|
Z50737-1-O |
Human Immunodeficiency Virus 3 (cluster #1 Of 1), Other |
Other |
40 |
0.55 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 3), Other |
Other |
9 |
0.59 |
Functional ≤ 10μM
|
|
Z80071-1-O |
CEM-0 (T-cell Leukemia) (cluster #1 Of 2), Other |
Other |
6 |
0.61 |
Functional ≤ 10μM
|
|
Z80073-1-O |
CEM-c113 (cluster #1 Of 1), Other |
Other |
6 |
0.61 |
Functional ≤ 10μM
|
|
Z80076-1-O |
CEM-SS (T-cell Leukemia) (cluster #1 Of 5), Other |
Other |
95 |
0.52 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
6180 |
0.38 |
Functional ≤ 10μM
|
|
Z80285-1-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
6 |
0.61 |
Functional ≤ 10μM
|
|
Z80294-1-O |
MT2 (Lymphocytes) (cluster #1 Of 1), Other |
Other |
6 |
0.61 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
10 |
0.59 |
Functional ≤ 10μM
|
|
Z80384-1-O |
PBL (Peripheral Blood Lymphocytes) (cluster #1 Of 1), Other |
Other |
29 |
0.56 |
Functional ≤ 10μM
|
|
Z80516-1-O |
SUP-T1 (cluster #1 Of 1), Other |
Other |
50 |
0.54 |
Functional ≤ 10μM |
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
40 |
0.55 |
Functional ≤ 10μM
|
|
Z80683-1-O |
ATH8 Cell Line (cluster #1 Of 1), Other |
Other |
400 |
0.47 |
Functional ≤ 10μM
|
|
Z80688-1-O |
B(EBV+) Cell Line (cluster #1 Of 1), Other |
Other |
1 |
0.66 |
Functional ≤ 10μM
|
|
Z80721-1-O |
BxT Cell Line (cluster #1 Of 1), Other |
Other |
55 |
0.53 |
Functional ≤ 10μM
|
|
Z80737-1-O |
C3H/3T3 (Embryonic Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
20 |
0.57 |
Functional ≤ 10μM
|
|
Z80760-1-O |
CFU-GM Cell Line (cluster #1 Of 1), Other |
Other |
1500 |
0.43 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
870 |
0.45 |
Functional ≤ 10μM
|
|
Z80897-1-O |
H9 (T-lymphoid Cells) (cluster #1 Of 2), Other |
Other |
9 |
0.59 |
Functional ≤ 10μM |
|
Z81072-2-O |
Jurkat (Acute Leukemic T-cells) (cluster #2 Of 10), Other |
Other |
10 |
0.59 |
Functional ≤ 10μM
|
|
Z81223-1-O |
M Cell Line (cluster #1 Of 1), Other |
Other |
15 |
0.58 |
Functional ≤ 10μM
|
|
Z81247-7-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #7 Of 9), Other |
Other |
9 |
0.59 |
Functional ≤ 10μM
|
|
Z80874-3-O |
CEM (T-cell Leukemia) (cluster #3 Of 4), Other |
Other |
700 |
0.45 |
ADME/T ≤ 10μM
|
|
Z80897-1-O |
H9 (T-lymphoid Cells) (cluster #1 Of 2), Other |
Other |
45 |
0.54 |
ADME/T ≤ 10μM
|
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
6 |
0.61 |
Binding ≤ 10μM
|
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
6 |
0.61 |
Binding ≤ 10μM
|
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 1), Viral |
Viruses |
8 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.10 |
-3.76 |
-14.65 |
2 |
9 |
0 |
134 |
267.245 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.32 |
4.71 |
-16.55 |
4 |
10 |
0 |
140 |
547.674 |
12 |
↓
|
|
|
|
Analogs
-
16952920
-
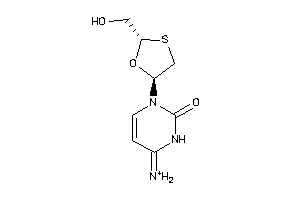
Draw
Identity
99%
90%
80%
70%
Vendors
And 56 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 4), Other |
Other |
27 |
0.71 |
Functional ≤ 10μM
|
|
Z101829-1-O |
HBV Genotype D (cluster #1 Of 1), Other |
Other |
460 |
0.59 |
Functional ≤ 10μM
|
|
Z50518-3-O |
Human Herpesvirus 4 (cluster #3 Of 5), Other |
Other |
400 |
0.60 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
2200 |
0.53 |
Functional ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
81 |
0.66 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
969 |
0.56 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
2000 |
0.53 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
60 |
0.67 |
Functional ≤ 10μM
|
|
Z50681-1-O |
Duck Hepatitis B Virus (cluster #1 Of 2), Other |
Other |
44 |
0.69 |
Functional ≤ 10μM
|
|
Z50755-1-O |
Anas Sp. (cluster #1 Of 1), Other |
Other |
100 |
0.65 |
Functional ≤ 10μM |
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
7000 |
0.48 |
Functional ≤ 10μM |
|
Z80947-1-O |
Hepatoblastoma Cell Line (cluster #1 Of 1), Other |
Other |
70 |
0.67 |
Functional ≤ 10μM
|
|
Z80949-1-O |
Hepatoma Cell Line (cluster #1 Of 1), Other |
Other |
10 |
0.75 |
Functional ≤ 10μM |
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 8), Other |
Other |
8 |
0.76 |
Functional ≤ 10μM
|
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
42 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.09 |
-2.17 |
-16.53 |
3 |
6 |
0 |
90 |
229.261 |
2 |
↓
|
|
|
|
Analogs
-
16025885
-
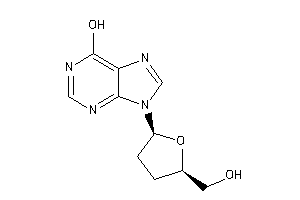
Draw
Identity
99%
90%
80%
70%
Vendors
And 47 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.95 |
1.07 |
-21.3 |
2 |
7 |
0 |
93 |
236.231 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.49 |
-0.99 |
-55.12 |
1 |
7 |
-1 |
96 |
235.223 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
1000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.10 |
-1.42 |
-16.23 |
1 |
7 |
0 |
94 |
370.814 |
6 |
↓
|
|
|
|
Analogs
-
3629271
-
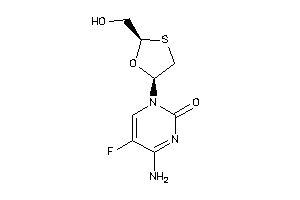
-
6524226
-
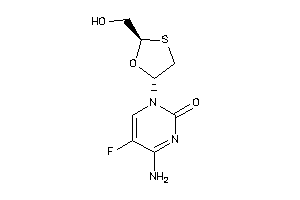
-
11616147
-
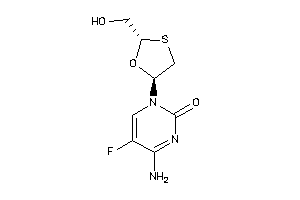
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
380 |
0.56 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
2100 |
0.50 |
Functional ≤ 10μM |
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 8), Other |
Other |
30 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.67 |
-6.62 |
-22.32 |
3 |
6 |
0 |
90 |
247.251 |
2 |
↓
|
|
|
|
Analogs
-
6524226
-

-
11616147
-

-
5472
-
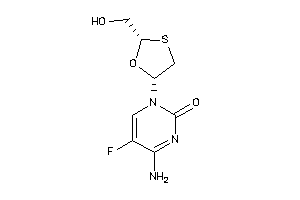
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.67 |
-2.06 |
-12.32 |
3 |
6 |
0 |
90 |
247.251 |
2 |
↓
|
|
|
|
Analogs
-
11616147
-

-
3629271
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
430 |
0.56 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
2100 |
0.50 |
Functional ≤ 10μM |
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 8), Other |
Other |
30 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.67 |
-6.58 |
-16.56 |
3 |
6 |
0 |
90 |
247.251 |
2 |
↓
|
|
|
|
Analogs
-
3629271
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
2100 |
0.50 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.67 |
-6.98 |
-12.71 |
3 |
6 |
0 |
90 |
247.251 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.16 |
Functional ≤ 10μM
|
|
CP2B6-3-E |
Cytochrome P450 2B6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.16 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.20 |
ADME/T ≤ 10μM
|
|
CP3A5-1-E |
Cytochrome P450 3A5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.19 |
ADME/T ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
2233 |
0.16 |
Functional ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
34 |
0.21 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
9664 |
0.14 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
25 |
0.21 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
90 |
0.20 |
Functional ≤ 10μM
|
|
Z50658-3-O |
Human Immunodeficiency Virus 2 (cluster #3 Of 4), Other |
Other |
84 |
0.20 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
38 |
0.21 |
Functional ≤ 10μM
|
|
Z80294-1-O |
MT2 (Lymphocytes) (cluster #1 Of 1), Other |
Other |
10 |
0.22 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
90 |
0.20 |
Functional ≤ 10μM |
|
Z102164-1-O |
Liver Microsomes (cluster #1 Of 1), Other |
Other |
40 |
0.21 |
ADME/T ≤ 10μM
|
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
60 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.51 |
12.91 |
-25.34 |
4 |
11 |
0 |
146 |
720.962 |
18 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
10.77 |
-65.17 |
1 |
6 |
-1 |
92 |
446.882 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.93 |
11.38 |
-50.45 |
3 |
7 |
1 |
74 |
352.466 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z101697-1-O |
Camelpox Virus (cluster #1 Of 1), Other |
Other |
5500 |
0.39 |
Functional ≤ 10μM
|
|
Z50515-2-O |
Human Herpesvirus 2 (cluster #2 Of 2), Other |
Other |
1100 |
0.44 |
Functional ≤ 10μM
|
|
Z50530-4-O |
Human Herpesvirus 5 (cluster #4 Of 5), Other |
Other |
90 |
0.52 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
90 |
0.52 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z80472-1-O |
SiHa (Cervical Squamous Cell Carcinoma) (cluster #1 Of 2), Other |
Other |
8970 |
0.37 |
Functional ≤ 10μM
|
|
Z80583-7-O |
Vero (Kidney Cells) (cluster #7 Of 7), Other |
Other |
1100 |
0.44 |
Functional ≤ 10μM
|
|
Z80874-7-O |
CEM (T-cell Leukemia) (cluster #7 Of 7), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z81247-5-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #5 Of 9), Other |
Other |
2160 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.14 |
-0.85 |
-60.55 |
4 |
10 |
-1 |
159 |
288.18 |
5 |
↓
|
|
|
|
Analogs
-
40147
-
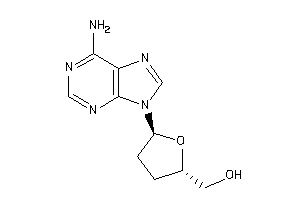
-
40148
-
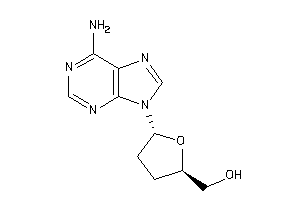
-
40149
-
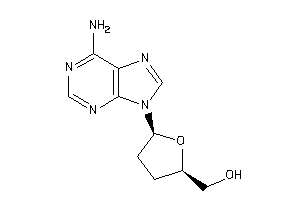
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.02 |
1.15 |
-10.79 |
3 |
7 |
0 |
99 |
235.247 |
2 |
↓
|
|
|
|
Analogs
-
5545870
-
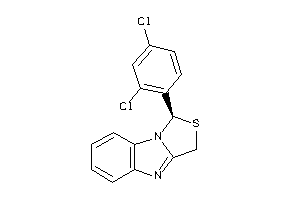
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
600 |
0.44 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
589 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.79 |
10.3 |
-8.77 |
0 |
2 |
0 |
18 |
321.232 |
1 |
↓
|
|
|
|
Analogs
-
3959130
-
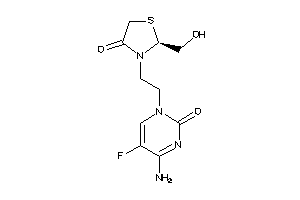
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50755-1-O |
Anas Sp. (cluster #1 Of 1), Other |
Other |
2000 |
0.42 |
Functional ≤ 10μM |
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.28 |
-6 |
-28.49 |
3 |
7 |
0 |
101 |
288.304 |
4 |
↓
|
|
|
|
Analogs
-
22752
-
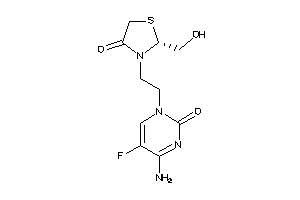
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50755-1-O |
Anas Sp. (cluster #1 Of 1), Other |
Other |
2000 |
0.42 |
Functional ≤ 10μM |
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.28 |
-6.03 |
-28.22 |
3 |
7 |
0 |
101 |
288.304 |
4 |
↓
|
|
|
|
Analogs
-
1725270
-
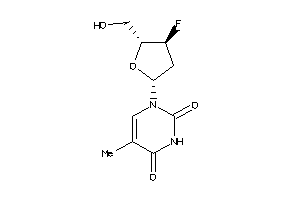
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
50 |
0.60 |
Binding ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
1 |
0.74 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.50 |
-0.36 |
-20.91 |
2 |
6 |
0 |
84 |
244.222 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
1.63 |
-10.91 |
2 |
6 |
0 |
84 |
308.359 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.72 |
-0.17 |
-52.66 |
1 |
6 |
-1 |
87 |
307.351 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.90 |
5.66 |
-11.87 |
1 |
5 |
0 |
64 |
302.374 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
5000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
0.74 |
-16.23 |
0 |
8 |
0 |
111 |
310.287 |
5 |
↓
|
|
|
|
Analogs
-
4093
-
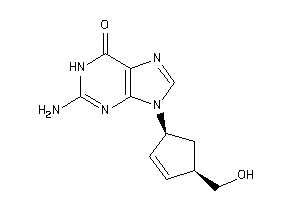
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.48 |
2.47 |
-17.8 |
4 |
7 |
0 |
110 |
247.258 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 50 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102020-1-O |
Porcine Endogenous Retrovirus (cluster #1 Of 1), Other |
Other |
7800 |
0.45 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
651 |
0.54 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
940 |
0.53 |
Functional ≤ 10μM
|
|
Z50612-1-O |
Moloney Murine Sarcoma Virus (cluster #1 Of 2), Other |
Other |
2500 |
0.49 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
790 |
0.53 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
770 |
0.53 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 3), Other |
Other |
80 |
0.62 |
Functional ≤ 10μM |
|
Z80071-1-O |
CEM-0 (T-cell Leukemia) (cluster #1 Of 2), Other |
Other |
800 |
0.53 |
Functional ≤ 10μM
|
|
Z80073-1-O |
CEM-c113 (cluster #1 Of 1), Other |
Other |
80 |
0.62 |
Functional ≤ 10μM
|
|
Z80076-1-O |
CEM-SS (T-cell Leukemia) (cluster #1 Of 5), Other |
Other |
59 |
0.63 |
Functional ≤ 10μM |
|
Z80077-1-O |
CEM-TK(-) (cluster #1 Of 1), Other |
Other |
10000 |
0.44 |
Functional ≤ 10μM |
|
Z80294-1-O |
MT2 (Lymphocytes) (cluster #1 Of 1), Other |
Other |
2000 |
0.50 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
710 |
0.54 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
770 |
0.53 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
90 |
0.62 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
270 |
0.57 |
Functional ≤ 10μM |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
500 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.54 |
-0.44 |
-16.04 |
2 |
6 |
0 |
84 |
224.216 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
500 |
0.55 |
Binding ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
651 |
0.54 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
770 |
0.53 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 3), Other |
Other |
80 |
0.62 |
Functional ≤ 10μM |
|
Z80071-1-O |
CEM-0 (T-cell Leukemia) (cluster #1 Of 2), Other |
Other |
800 |
0.53 |
Functional ≤ 10μM
|
|
Z80073-1-O |
CEM-c113 (cluster #1 Of 1), Other |
Other |
80 |
0.62 |
Functional ≤ 10μM
|
|
Z80076-1-O |
CEM-SS (T-cell Leukemia) (cluster #1 Of 5), Other |
Other |
59 |
0.63 |
Functional ≤ 10μM |
|
Z80077-1-O |
CEM-TK(-) (cluster #1 Of 1), Other |
Other |
10000 |
0.44 |
Functional ≤ 10μM |
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
770 |
0.53 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
270 |
0.57 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.54 |
-1.51 |
-9.78 |
2 |
6 |
0 |
84 |
224.216 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
500 |
0.55 |
Binding ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
651 |
0.54 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
770 |
0.53 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 3), Other |
Other |
80 |
0.62 |
Functional ≤ 10μM |
|
Z80071-1-O |
CEM-0 (T-cell Leukemia) (cluster #1 Of 2), Other |
Other |
800 |
0.53 |
Functional ≤ 10μM
|
|
Z80073-1-O |
CEM-c113 (cluster #1 Of 1), Other |
Other |
80 |
0.62 |
Functional ≤ 10μM
|
|
Z80076-1-O |
CEM-SS (T-cell Leukemia) (cluster #1 Of 5), Other |
Other |
59 |
0.63 |
Functional ≤ 10μM |
|
Z80077-1-O |
CEM-TK(-) (cluster #1 Of 1), Other |
Other |
10000 |
0.44 |
Functional ≤ 10μM |
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
770 |
0.53 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
270 |
0.57 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.54 |
-0.47 |
-16.99 |
2 |
6 |
0 |
84 |
224.216 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
651 |
0.54 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
770 |
0.53 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 3), Other |
Other |
80 |
0.62 |
Functional ≤ 10μM |
|
Z80071-1-O |
CEM-0 (T-cell Leukemia) (cluster #1 Of 2), Other |
Other |
800 |
0.53 |
Functional ≤ 10μM
|
|
Z80073-1-O |
CEM-c113 (cluster #1 Of 1), Other |
Other |
80 |
0.62 |
Functional ≤ 10μM
|
|
Z80076-1-O |
CEM-SS (T-cell Leukemia) (cluster #1 Of 5), Other |
Other |
59 |
0.63 |
Functional ≤ 10μM |
|
Z80077-1-O |
CEM-TK(-) (cluster #1 Of 1), Other |
Other |
10000 |
0.44 |
Functional ≤ 10μM |
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
770 |
0.53 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
270 |
0.57 |
Functional ≤ 10μM |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
500 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.54 |
0 |
-17.97 |
2 |
6 |
0 |
84 |
224.216 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.76 |
0.84 |
-10.49 |
0 |
2 |
0 |
17 |
288.322 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.35 |
0.99 |
-27.95 |
1 |
2 |
1 |
19 |
289.33 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
9000 |
0.35 |
Binding ≤ 10μM
|
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
101 |
0.49 |
Binding ≤ 10μM
|
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 1), Viral |
Viruses |
52 |
0.51 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
9900 |
0.35 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
50 |
0.51 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 3), Other |
Other |
40 |
0.52 |
Functional ≤ 10μM
|
|
Z80294-1-O |
MT2 (Lymphocytes) (cluster #1 Of 1), Other |
Other |
303 |
0.46 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
96 |
0.49 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
84 |
0.50 |
Functional ≤ 10μM
|
|
Z80897-1-O |
H9 (T-lymphoid Cells) (cluster #1 Of 2), Other |
Other |
5000 |
0.37 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
89 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
6.59 |
-11.74 |
1 |
5 |
0 |
64 |
266.304 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.39 |
7.06 |
-37.11 |
2 |
5 |
1 |
65 |
267.312 |
1 |
↓
|
|
|
|
Analogs
-
33822027
-
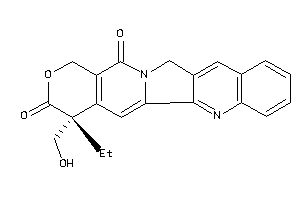
-
33822028
-
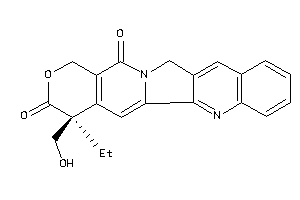
-
1087
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 55 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.31 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.27 |
Binding ≤ 10μM
|
|
CHK1-1-E |
Serine/threonine-protein Kinase Chk1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR1-1-E |
Somatostatin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR2-1-E |
Somatostatin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR3-1-E |
Somatostatin Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR4-1-E |
Somatostatin Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR5-1-E |
Somatostatin Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.40 |
Functional ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.40 |
Functional ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.40 |
Functional ≤ 10μM
|
|
Z100501-1-O |
Lu1 (Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
29 |
0.41 |
Functional ≤ 10μM
|
|
Z100733-1-O |
CT26 (Colon Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
34 |
0.40 |
Functional ≤ 10μM
|
|
Z100752-1-O |
Renca (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
304 |
0.35 |
Functional ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
11 |
0.43 |
Functional ≤ 10μM
|
|
Z103408-1-O |
SW626 (cluster #1 Of 2), Other |
Other |
29 |
0.41 |
Functional ≤ 10μM
|
|
Z103413-1-O |
SW948 (cluster #1 Of 1), Other |
Other |
160 |
0.37 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
794 |
0.33 |
Functional ≤ 10μM
|
|
Z80012-1-O |
A 172 (Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
29 |
0.41 |
Functional ≤ 10μM
|
|
Z80012-1-O |
A 172 (Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
140 |
0.37 |
Functional ≤ 10μM |
|
Z80019-1-O |
A-427 (Lung Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
24 |
0.41 |
Functional ≤ 10μM
|
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
20 |
0.41 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
9550 |
0.27 |
Functional ≤ 10μM |
|
Z80056-1-O |
CAKI-2 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
3960 |
0.29 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
5300 |
0.28 |
Functional ≤ 10μM
|
|
Z80068-3-O |
CCRF-SB (Lymphoblastic Leukemia Cells) (cluster #3 Of 5), Other |
Other |
3 |
0.46 |
Functional ≤ 10μM
|
|
Z80099-1-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
5 |
0.45 |
Functional ≤ 10μM
|
|
Z80115-1-O |
DC3F (cluster #1 Of 1), Other |
Other |
6 |
0.44 |
Functional ≤ 10μM |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
102 |
0.38 |
Functional ≤ 10μM
|
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
210 |
0.36 |
Functional ≤ 10μM |
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
940 |
0.32 |
Functional ≤ 10μM
|
|
Z80145-3-O |
H69 (cluster #3 Of 4), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
7 |
0.44 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
89 |
0.38 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
80 |
0.38 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
90 |
0.38 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
80 |
0.38 |
Functional ≤ 10μM
|
|
Z80188-1-O |
KB 3-1 (Cervical Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
20 |
0.41 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
35 |
0.40 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
60 |
0.39 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
3100 |
0.30 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
7420 |
0.28 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
17 |
0.42 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
4 |
0.45 |
Functional ≤ 10μM
|
|
Z80316-1-O |
NCI-H128 (cluster #1 Of 1), Other |
Other |
18 |
0.42 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z80372-1-O |
P388CPT5 (cluster #1 Of 1), Other |
Other |
2800 |
0.30 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
57 |
0.39 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
1000 |
0.32 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
40 |
0.40 |
Functional ≤ 10μM |
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
20 |
0.41 |
Functional ≤ 10μM
|
|
Z80482-2-O |
SK-MEL-2 (Melanoma Cells) (cluster #2 Of 4), Other |
Other |
7860 |
0.27 |
Functional ≤ 10μM |
|
Z80485-1-O |
SK-MEL-28 (Melanoma Cells) (cluster #1 Of 6), Other |
Other |
40 |
0.40 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
51 |
0.39 |
Functional ≤ 10μM
|
|
Z80495-1-O |
SK-VLB (cluster #1 Of 1), Other |
Other |
53 |
0.39 |
Functional ≤ 10μM
|
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
88 |
0.38 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
600 |
0.34 |
Functional ≤ 10μM
|
|
Z80623-1-O |
Panel (56 Tumour Cell Lines) (cluster #1 Of 1), Other |
Other |
40 |
0.40 |
Functional ≤ 10μM
|
|
Z80647-1-O |
833K Cell Line (cluster #1 Of 1), Other |
Other |
10 |
0.43 |
Functional ≤ 10μM |
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
91 |
0.38 |
Functional ≤ 10μM |
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
7 |
0.44 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
207 |
0.36 |
Functional ≤ 10μM
|
|
Z80751-1-O |
Caov-3 Cell Line (cluster #1 Of 1), Other |
Other |
30 |
0.41 |
Functional ≤ 10μM |
|
Z80751-1-O |
Caov-3 Cell Line (cluster #1 Of 1), Other |
Other |
32 |
0.40 |
Functional ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
45 |
0.40 |
Functional ≤ 10μM
|
|
Z80799-1-O |
RPMI 8402 (Pre-T-lymphoblastoid Cells) (cluster #1 Of 1), Other |
Other |
6 |
0.44 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80898-1-O |
NCI-H928 (cluster #1 Of 1), Other |
Other |
8 |
0.44 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z80930-1-O |
HEC-1B Cell Line (cluster #1 Of 1), Other |
Other |
60 |
0.39 |
Functional ≤ 10μM
|
|
Z80930-1-O |
HEC-1B Cell Line (cluster #1 Of 1), Other |
Other |
8640 |
0.27 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
17 |
0.42 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
60 |
0.39 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
330 |
0.35 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
7 |
0.44 |
Functional ≤ 10μM
|
|
Z81054-1-O |
SF-268 (Glioblastoma Cells) (cluster #1 Of 3), Other |
Other |
59 |
0.39 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
6 |
0.44 |
Functional ≤ 10μM
|
|
Z81080-1-O |
KATO-III Cell Line (cluster #1 Of 1), Other |
Other |
448 |
0.34 |
Functional ≤ 10μM
|
|
Z81080-1-O |
KATO-III Cell Line (cluster #1 Of 1), Other |
Other |
1170 |
0.32 |
Functional ≤ 10μM |
|
Z81084-1-O |
KB VCR R (cluster #1 Of 1), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z81134-1-O |
L2987 (Lung Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
400 |
0.34 |
Functional ≤ 10μM
|
|
Z81160-1-O |
Lewis Lung Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
33 |
0.40 |
Functional ≤ 10μM
|
|
Z81164-1-O |
LL Cell Line (cluster #1 Of 2), Other |
Other |
33 |
0.40 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
86 |
0.38 |
Functional ≤ 10μM
|
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
48 |
0.39 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
710 |
0.33 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
940 |
0.32 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
970 |
0.32 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
2 |
0.47 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
83 |
0.38 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
890 |
0.33 |
ADME/T ≤ 10μM |
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
500 |
0.34 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
6.76 |
-12.81 |
1 |
6 |
0 |
81 |
348.358 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
1800 |
0.32 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
1800 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.67 |
-1.59 |
-11.79 |
2 |
6 |
0 |
91 |
378.837 |
5 |
↓
|
|
|
|
Analogs
-
5844601
-
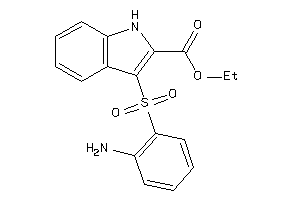
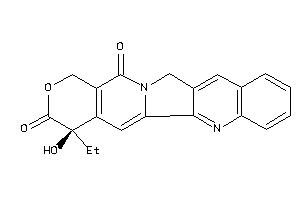
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
1900 |
0.31 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
1900 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
-3.68 |
-13.6 |
3 |
6 |
0 |
102 |
413.282 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-3-O |
Human Immunodeficiency Virus 1 (cluster #3 Of 10), Other |
Other |
2500 |
0.33 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
2500 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.25 |
-1.47 |
-8.21 |
3 |
4 |
0 |
68 |
381.284 |
5 |
↓
|
|
|
|
Analogs
-
39908
-
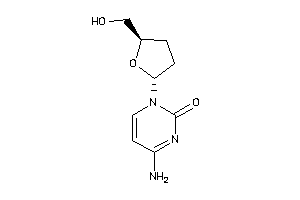
-
17175067
-
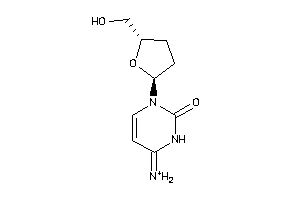
-
17175068
-
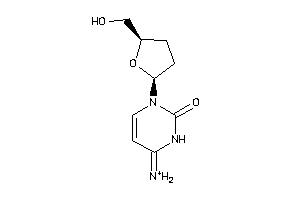
Draw
Identity
99%
90%
80%
70%
Vendors
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR4-1-E |
C-X-C Chemokine Receptor Type 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
88 |
0.66 |
Functional ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
2300 |
0.53 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
910 |
0.56 |
Functional ≤ 10μM
|
|
Z50624-1-O |
Human Adenovirus Type 2 (cluster #1 Of 1), Other |
Other |
3300 |
0.51 |
Functional ≤ 10μM
|
|
Z50658-2-O |
Human Immunodeficiency Virus 2 (cluster #2 Of 4), Other |
Other |
690 |
0.58 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
800 |
0.57 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 3), Other |
Other |
125 |
0.64 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
22 |
0.71 |
Functional ≤ 10μM
|
|
Z80076-1-O |
CEM-SS (T-cell Leukemia) (cluster #1 Of 5), Other |
Other |
50 |
0.68 |
Functional ≤ 10μM
|
|
Z80294-1-O |
MT2 (Lymphocytes) (cluster #1 Of 1), Other |
Other |
994 |
0.56 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
300 |
0.61 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
70 |
0.67 |
Functional ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 8), Other |
Other |
3200 |
0.51 |
Functional ≤ 10μM |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
51 |
0.68 |
Binding ≤ 10μM
|
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 1), Viral |
Viruses |
31 |
0.70 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.06 |
-2.38 |
-18.62 |
3 |
6 |
0 |
90 |
211.221 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
10.58 |
-21.49 |
2 |
13 |
0 |
167 |
501.477 |
15 |
↓
|
|