|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
1800 |
0.29 |
Functional ≤ 10μM |
|
Z80224-7-O |
MCF7 (Breast Carcinoma Cells) (cluster #7 Of 14), Other |
Other |
1900 |
0.29 |
Functional ≤ 10μM |
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
2400 |
0.28 |
Functional ≤ 10μM |
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
3900 |
0.27 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
5.94 |
-9.99 |
2 |
6 |
0 |
93 |
390.476 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3S,5R,8R,9S,10S,13R,14S,16S,17R)-14-hydroxy-3-[(2R,4R,5S,6R)-5-hydroxy-4-methoxy-6-methyl-tetrahyd
[(3S,5R,8R,9S,10S,13R,14S,16S,17…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
510 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
9.72 |
-17.67 |
2 |
9 |
0 |
121 |
576.727 |
6 |
↓
|
|
|
|
Analogs
-
67911417
-
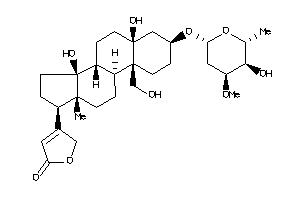
-
70691688
-
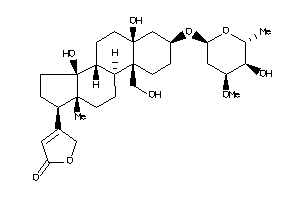
-
70691854
-
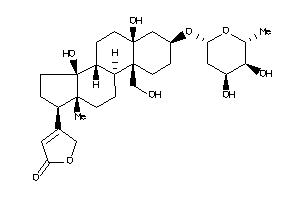
-
70708576
-
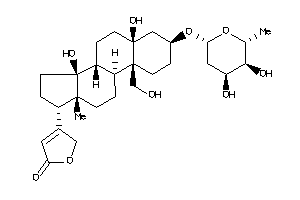
-
36372256
-
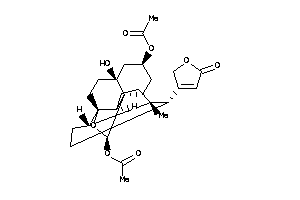
Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[(3S,5R,8R,9S,10S,13R,14S,17R)-14-hydroxy-3-[(2R,4R,5S,6R)-5-hydroxy-4-methoxy-6-methyl-tetrahydro
3-[(3S,5R,8R,9S,10S,13R,14S,17R)…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
480 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
8.78 |
-16.05 |
2 |
7 |
0 |
94 |
518.691 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
15449189
-
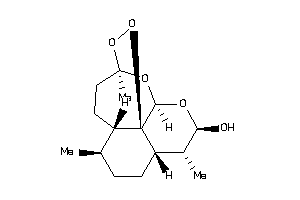
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
56 |
0.51 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
6 |
0.58 |
Functional ≤ 10μM
|
|
Z50436-3-O |
Filobasidiella Neoformans (cluster #3 Of 3), Other |
Other |
280 |
0.46 |
Functional ≤ 10μM
|
|
Z50442-2-O |
Candida Albicans (cluster #2 Of 4), Other |
Other |
880 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-3-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #3 Of 12), Other |
Other |
2900 |
0.39 |
Functional ≤ 10μM
|
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
750 |
0.43 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
850 |
0.42 |
Functional ≤ 10μM
|
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
540 |
0.44 |
Functional ≤ 10μM
|
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
630 |
0.43 |
Functional ≤ 10μM
|
|
Z81057-2-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #2 Of 4), Other |
Other |
8910 |
0.35 |
Functional ≤ 10μM
|
|
Z81072-2-O |
Jurkat (Acute Leukemic T-cells) (cluster #2 Of 10), Other |
Other |
650 |
0.43 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
460 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
3.29 |
-6.48 |
1 |
5 |
0 |
57 |
284.352 |
0 |
↓
|
|
|
|
Analogs
-
15449187
-
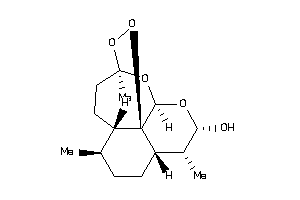
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
56 |
0.51 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
6 |
0.58 |
Functional ≤ 10μM
|
|
Z50436-3-O |
Filobasidiella Neoformans (cluster #3 Of 3), Other |
Other |
280 |
0.46 |
Functional ≤ 10μM
|
|
Z50442-2-O |
Candida Albicans (cluster #2 Of 4), Other |
Other |
880 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-3-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #3 Of 12), Other |
Other |
2900 |
0.39 |
Functional ≤ 10μM
|
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
750 |
0.43 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
850 |
0.42 |
Functional ≤ 10μM
|
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
540 |
0.44 |
Functional ≤ 10μM
|
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
630 |
0.43 |
Functional ≤ 10μM
|
|
Z81057-2-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #2 Of 4), Other |
Other |
8910 |
0.35 |
Functional ≤ 10μM
|
|
Z81072-2-O |
Jurkat (Acute Leukemic T-cells) (cluster #2 Of 10), Other |
Other |
650 |
0.43 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
460 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
3.66 |
-6.09 |
1 |
5 |
0 |
57 |
284.352 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-10-O |
MCF7 (Breast Carcinoma Cells) (cluster #10 Of 14), Other |
Other |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
10000 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.72 |
20.05 |
-49.34 |
0 |
6 |
0 |
68 |
525.777 |
26 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-10-O |
MCF7 (Breast Carcinoma Cells) (cluster #10 Of 14), Other |
Other |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
10000 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-10-O |
MCF7 (Breast Carcinoma Cells) (cluster #10 Of 14), Other |
Other |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
10000 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.72 |
19.79 |
-46.4 |
0 |
6 |
0 |
68 |
525.777 |
26 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80224-10-O |
MCF7 (Breast Carcinoma Cells) (cluster #10 Of 14), Other |
Other |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
10000 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
36532609
-
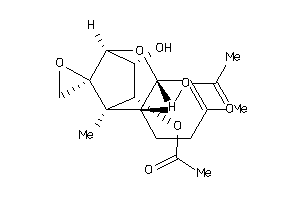
-
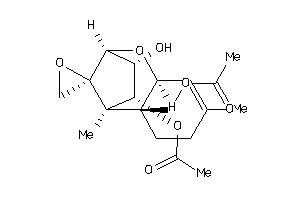
36532611
-
-
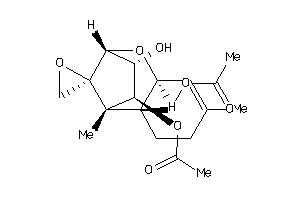
38495208
-
-
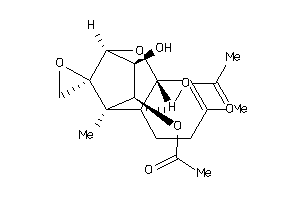
5053041
-
-
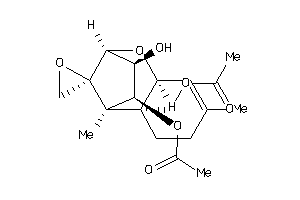
5053042
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80021-3-O |
A498 (Renal Carcinoma Cells) (cluster #3 Of 5), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80099-1-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80186-4-O |
K562 (Erythroleukemia Cells) (cluster #4 Of 11), Other |
Other |
3 |
0.46 |
Functional ≤ 10μM
|
|
Z80214-2-O |
M14 (Melanoma Cells) (cluster #2 Of 3), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80224-2-O |
MCF7 (Breast Carcinoma Cells) (cluster #2 Of 14), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80285-1-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80438-1-O |
RXF 393 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80570-1-O |
UACC-62 (Melanoma Cells) (cluster #1 Of 3), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80640-1-O |
786-0 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z81245-4-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #4 Of 6), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z81246-1-O |
MDA-N (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z81280-3-O |
NCI-H226 (Non-small Cell Lung Carcinoma Cells) (cluster #3 Of 3), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z81283-2-O |
NCI-H522 (Non-small Cell Lung Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.54 |
4.92 |
-12.41 |
1 |
7 |
0 |
95 |
366.41 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
-5.86 |
-18.18 |
3 |
9 |
0 |
131 |
532.63 |
2 |
↓
|
|
|
|
Analogs
-
43427402
-
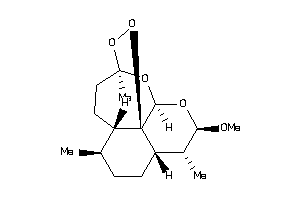
-
8214360
-
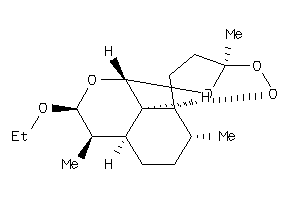
-
8214363
-
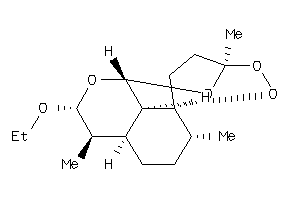
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
5.25 |
-6.37 |
0 |
5 |
0 |
46 |
298.379 |
1 |
↓
|
|
|
|
Analogs
-
6361655
-
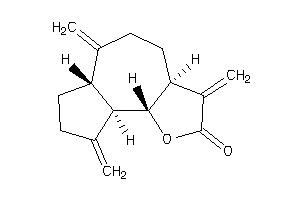
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
10000 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
3.13 |
-7.99 |
0 |
2 |
0 |
26 |
230.307 |
0 |
↓
|
|
|
|
Analogs
-
5360089
-
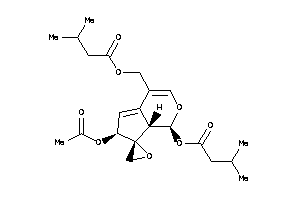
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
3700 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.31 |
4.07 |
-15.4 |
0 |
8 |
0 |
100 |
422.474 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.33 |
4.91 |
-22.61 |
1 |
6 |
0 |
82 |
360.406 |
0 |
↓
|
|