|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 68 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100081-3-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #3 Of 4), Other |
Other |
150 |
0.96 |
Functional ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
2000 |
0.80 |
Functional ≤ 10μM
|
|
Z80064-3-O |
CCRF-CEM (T-cell Leukemia) (cluster #3 Of 9), Other |
Other |
1000 |
0.84 |
Functional ≤ 10μM
|
|
Z80068-2-O |
CCRF-SB (Lymphoblastic Leukemia Cells) (cluster #2 Of 5), Other |
Other |
1000 |
0.84 |
Functional ≤ 10μM
|
|
Z80125-5-O |
DU-145 (Prostate Carcinoma) (cluster #5 Of 9), Other |
Other |
2000 |
0.80 |
Functional ≤ 10μM
|
|
Z80193-6-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #6 Of 12), Other |
Other |
24 |
1.07 |
Functional ≤ 10μM
|
|
Z80224-4-O |
MCF7 (Breast Carcinoma Cells) (cluster #4 Of 14), Other |
Other |
5800 |
0.73 |
Functional ≤ 10μM
|
|
Z80295-4-O |
MT4 (Lymphocytes) (cluster #4 Of 8), Other |
Other |
100 |
0.98 |
Functional ≤ 10μM
|
|
Z80936-3-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #3 Of 4), Other |
Other |
7 |
1.14 |
Functional ≤ 10μM
|
|
Z81020-5-O |
HepG2 (Hepatoblastoma Cells) (cluster #5 Of 8), Other |
Other |
8000 |
0.71 |
Functional ≤ 10μM
|
|
Z81247-5-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #5 Of 9), Other |
Other |
4100 |
0.75 |
Functional ≤ 10μM
|
|
Z80178-1-O |
J774.A1 (Macrophage Cells) (cluster #1 Of 2), Other |
Other |
3 |
1.19 |
ADME/T ≤ 10μM
|
|
Z80936-3-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #3 Of 4), Other |
Other |
7 |
1.14 |
ADME/T ≤ 10μM
|
|
Z81244-2-O |
J774 (Macrophage Cells) (cluster #2 Of 3), Other |
Other |
3 |
1.19 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.39 |
4.06 |
-18.68 |
2 |
4 |
0 |
57 |
152.182 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.34 |
3.67 |
-44.86 |
1 |
4 |
-1 |
54 |
151.174 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.34 |
3.66 |
-43.68 |
1 |
4 |
-1 |
54 |
151.174 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
2.88 |
-8.41 |
2 |
4 |
0 |
69 |
212.208 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
0.64 |
-10.62 |
0 |
3 |
0 |
43 |
247.253 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 84 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z101794-1-O |
Felid Herpesvirus 1 (cluster #1 Of 1), Other |
Other |
1000 |
0.53 |
Functional ≤ 10μM |
|
Z50515-2-O |
Human Herpesvirus 2 (cluster #2 Of 2), Other |
Other |
9600 |
0.44 |
Functional ≤ 10μM
|
|
Z50517-1-O |
Human Herpesvirus 5 Strain AD169 (cluster #1 Of 2), Other |
Other |
7600 |
0.45 |
Functional ≤ 10μM
|
|
Z50518-3-O |
Human Herpesvirus 4 (cluster #3 Of 5), Other |
Other |
6900 |
0.45 |
Functional ≤ 10μM
|
|
Z50527-1-O |
Human Herpesvirus 3 (cluster #1 Of 3), Other |
Other |
900 |
0.53 |
Functional ≤ 10μM
|
|
Z50527-3-O |
Human Herpesvirus 3 (cluster #3 Of 3), Other |
Other |
4900 |
0.46 |
Functional ≤ 10μM |
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
7500 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
90 |
0.62 |
Functional ≤ 10μM
|
|
Z50600-1-O |
Vaccinia Virus (cluster #1 Of 2), Other |
Other |
384 |
0.56 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
990 |
0.53 |
Functional ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
20 |
0.67 |
Functional ≤ 10μM
|
|
Z80040-1-O |
BHK-21 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.50 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
5000 |
0.46 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
2900 |
0.48 |
Functional ≤ 10μM
|
|
Z80583-7-O |
Vero (Kidney Cells) (cluster #7 Of 7), Other |
Other |
7540 |
0.45 |
Functional ≤ 10μM
|
|
Z80942-1-O |
HEL (Embryonic Lung Cells) (cluster #1 Of 2), Other |
Other |
900 |
0.53 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 4), Other |
Other |
8100 |
0.45 |
Functional ≤ 10μM
|
|
Z81247-5-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #5 Of 9), Other |
Other |
8710 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.61 |
-0.6 |
-19.69 |
4 |
8 |
0 |
119 |
225.208 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z101697-1-O |
Camelpox Virus (cluster #1 Of 1), Other |
Other |
5500 |
0.39 |
Functional ≤ 10μM
|
|
Z50515-2-O |
Human Herpesvirus 2 (cluster #2 Of 2), Other |
Other |
1100 |
0.44 |
Functional ≤ 10μM
|
|
Z50530-4-O |
Human Herpesvirus 5 (cluster #4 Of 5), Other |
Other |
90 |
0.52 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
90 |
0.52 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z80472-1-O |
SiHa (Cervical Squamous Cell Carcinoma) (cluster #1 Of 2), Other |
Other |
8970 |
0.37 |
Functional ≤ 10μM
|
|
Z80583-7-O |
Vero (Kidney Cells) (cluster #7 Of 7), Other |
Other |
1100 |
0.44 |
Functional ≤ 10μM
|
|
Z80874-7-O |
CEM (T-cell Leukemia) (cluster #7 Of 7), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
|
Z81247-5-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #5 Of 9), Other |
Other |
2160 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.14 |
-0.85 |
-60.55 |
4 |
10 |
-1 |
159 |
288.18 |
5 |
↓
|
|
|
|
Analogs
-
4541062
-
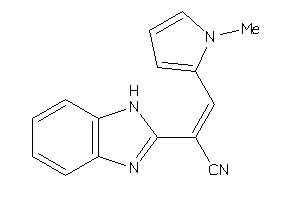
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81247-5-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #5 Of 9), Other |
Other |
800 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
7.84 |
-10.23 |
1 |
4 |
0 |
57 |
248.289 |
2 |
↓
|
|
|
|
Analogs
-
15223702
-
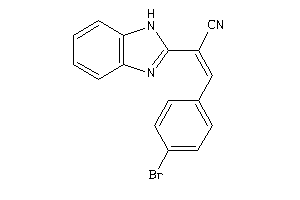
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81247-5-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #5 Of 9), Other |
Other |
9000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.19 |
0.07 |
-7.94 |
1 |
3 |
0 |
52 |
324.181 |
2 |
↓
|
|
|
|
Analogs
-
5635513
-
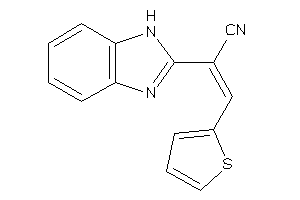
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
7.54 |
-7.46 |
1 |
3 |
0 |
52 |
251.314 |
2 |
↓
|
|