|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9R,10aR)-6,6-dimethyl-3-BLAHyl-6a,7,8,9,10,10a-hexahydrobenzo[c]chromene-1,9-diol
(6aR,9R,10aR)-6,6-dimethyl-3-BLA…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1800 |
0.29 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
4.63 |
-8.23 |
2 |
5 |
0 |
62 |
385.504 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.58 |
4.37 |
-34.66 |
3 |
5 |
1 |
63 |
386.512 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9S,10aR)-6,6-dimethyl-3-(BLAHylmethyl)-6a,7,8,9,10,10a-hexahydrobenzo[c]chromene-1,9-diol
(6aR,9S,10aR)-6,6-dimethyl-3-(BL…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.28 |
5.41 |
-37.18 |
3 |
5 |
1 |
63 |
400.539 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.28 |
3.5 |
-6.47 |
2 |
5 |
0 |
62 |
399.531 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9S,10aR)-6,6-dimethyl-3-BLAHyl-6a,7,8,9,10,10a-hexahydrobenzo[c]chromene-1,9-diol
(6aR,9S,10aR)-6,6-dimethyl-3-BLA…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
4.77 |
-6.59 |
2 |
5 |
0 |
62 |
385.504 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.58 |
4.52 |
-32.76 |
3 |
5 |
1 |
63 |
386.512 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9R,10aR)-6,6-dimethyl-3-(BLAHylmethyl)-6a,7,8,9,10,10a-hexahydrobenzo[c]chromene-1,9-diol
(6aR,9R,10aR)-6,6-dimethyl-3-(BL…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.24 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8700 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.28 |
5.26 |
-38.7 |
3 |
5 |
1 |
63 |
400.539 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.28 |
3.35 |
-8.38 |
2 |
5 |
0 |
62 |
399.531 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9R,10aR)-N-(1-adamantyl)-1,9-dihydroxy-6,6-dimethyl-6a,7,8,9,10,10a-hexahydrobenzo[c]chromene-3
(6aR,9R,10aR)-N-(1-adamantyl)-1,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.00 |
5.87 |
-13.12 |
3 |
5 |
0 |
79 |
425.569 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(6aR,9R,10aR)-1,9-dihydroxy-6,6-dimethyl-6a,7,8,9,10,10a-hexahydrobenzo[c]chromen-3-yl]-BLAHyl-meth
[(6aR,9R,10aR)-1,9-dihydroxy-6,6…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2500 |
0.26 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR2_MOUSE |
P47936
|
Cannabinoid CB2 Receptor, Mouse |
2500 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
4.17 |
-14.04 |
2 |
6 |
0 |
79 |
413.514 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9S,10aR)-N-(1-adamantyl)-1,9-dihydroxy-6,6-dimethyl-6a,7,8,9,10,10a-hexahydrobenzo[c]chromene-3
(6aR,9S,10aR)-N-(1-adamantyl)-1,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.23 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR2_MOUSE |
P47936
|
Cannabinoid CB2 Receptor, Mouse |
7500 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.00 |
6.02 |
-11.32 |
3 |
5 |
0 |
79 |
425.569 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(6aR,9S,10aR)-1,9-dihydroxy-6,6-dimethyl-6a,7,8,9,10,10a-hexahydrobenzo[c]chromen-3-yl]-BLAHyl-meth
[(6aR,9S,10aR)-1,9-dihydroxy-6,6…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8600 |
0.24 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR2_MOUSE |
P47936
|
Cannabinoid CB2 Receptor, Mouse |
8600 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
4.32 |
-11.79 |
2 |
6 |
0 |
79 |
413.514 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9R,10aR)-6,6-dimethyl-3-BLAHyl-6a,7,8,9,10,10a-hexahydrobenzo[c]chromene-1,9-diol
(6aR,9R,10aR)-6,6-dimethyl-3-BLA…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
23 |
0.38 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.92 |
5.95 |
-9.12 |
2 |
4 |
0 |
59 |
384.516 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9R,10aR)-3-[(2-adamantylamino)methyl]-6,6-dimethyl-6a,7,8,9,10,10a-hexahydrobenzo[c]chromene-1,
(6aR,9R,10aR)-3-[(2-adamantylami…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6200 |
0.24 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR2_MOUSE |
P47936
|
Cannabinoid CB2 Receptor, Mouse |
6200 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.21 |
7.5 |
-43.66 |
4 |
4 |
1 |
66 |
412.594 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9R,10aR)-N-(2-adamantyl)-1,9-dihydroxy-6,6-dimethyl-6a,7,8,9,10,10a-hexahydrobenzo[c]chromene-3
(6aR,9R,10aR)-N-(2-adamantyl)-1,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.84 |
6.39 |
-13.25 |
3 |
5 |
0 |
79 |
425.569 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.36 |
12.54 |
-5.72 |
1 |
2 |
0 |
29 |
378.556 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.36 |
12.54 |
-5.58 |
1 |
2 |
0 |
29 |
378.556 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.36 |
12.6 |
-5.59 |
1 |
2 |
0 |
29 |
378.556 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.36 |
12.6 |
-5.51 |
1 |
2 |
0 |
29 |
378.556 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,10S,10aS)-6,6-dimethyl-3-[(1S)-1-methyl-4-phenyl-butyl]-6a,7,8,9,10,10a-hexahydrobenzo[c]isochr
(6aR,10S,10aS)-6,6-dimethyl-3-[(…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.21 |
9.93 |
-7.77 |
2 |
3 |
0 |
50 |
394.555 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,10S,10aS)-6,6-dimethyl-3-[(1R)-1-methyl-4-phenyl-butyl]-6a,7,8,9,10,10a-hexahydrobenzo[c]isochr
(6aR,10S,10aS)-6,6-dimethyl-3-[(…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.21 |
9.93 |
-7.67 |
2 |
3 |
0 |
50 |
394.555 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,10S,10aR)-6,6-dimethyl-3-[(1S)-1-methyl-4-phenyl-butyl]-6a,7,8,9,10,10a-hexahydrobenzo[c]isochr
(6aR,10S,10aR)-6,6-dimethyl-3-[(…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.21 |
9.74 |
-7.52 |
2 |
3 |
0 |
50 |
394.555 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,10S,10aR)-6,6-dimethyl-3-[(1R)-1-methyl-4-phenyl-butyl]-6a,7,8,9,10,10a-hexahydrobenzo[c]isochr
(6aR,10S,10aR)-6,6-dimethyl-3-[(…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.21 |
9.74 |
-7.47 |
2 |
3 |
0 |
50 |
394.555 |
5 |
↓
|
|
|
|
Analogs
-
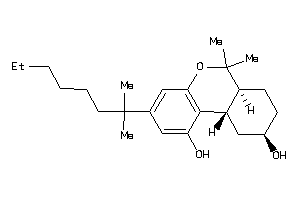
4212312
-
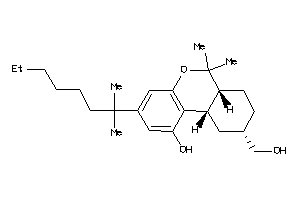
-
26176150
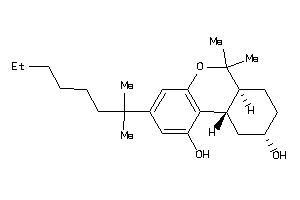
-
-
31983344
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9S,10aR)-3-(1,1-dimethylheptyl)-6,6-dimethyl-9-methylol-6a,7,8,9,10,10a-hexahydrobenzo[c]isochr
(6aR,9S,10aR)-3-(1,1-dimethylhep…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.37 |
8.83 |
-5.29 |
2 |
3 |
0 |
50 |
388.592 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9S,10aR)-3-[1-(5-bromopentyl)cyclobutyl]-6,6-dimethyl-6a,7,8,9,10,10a-hexahydrobenzo[c]chromene
(6aR,9S,10aR)-3-[1-(5-bromopenty…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.88 |
9.26 |
-6.45 |
2 |
3 |
0 |
50 |
451.445 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9R,10aR)-3-(1-hexylcyclobutyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydrobenzo[c]isochromene-1,9-di
(6aR,9R,10aR)-3-(1-hexylcyclobut…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.54 |
9.17 |
-7.15 |
2 |
3 |
0 |
50 |
386.576 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9R,10aR)-3-(2-hexyl-1,3-dithiolan-2-yl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydrobenzo[c]isochrome
(6aR,9R,10aR)-3-(2-hexyl-1,3-dit…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.40 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
14 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.76 |
9.55 |
-9.29 |
2 |
3 |
0 |
50 |
436.683 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6aR,9R,10aR)-3-[1-(5-bromopentyl)cyclobutyl]-6,6-dimethyl-6a,7,8,9,10,10a-hexahydrobenzo[c]chromene
(6aR,9R,10aR)-3-[1-(5-bromopenty…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.88 |
9.11 |
-8.16 |
2 |
3 |
0 |
50 |
451.445 |
6 |
↓
|
|