Substance Information
| In ZINC since |
Heavy atoms |
Benign functionality |
| June 21st, 2011 |
28 |
Yes
|
Popular Name:
(6aR,9R,10aR)-3-(1-hexylcyclobutyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydrobenzo[c]isochromene-1,9-di
(6aR,9R,10aR)-3-(1-hexylcyclobut…
Find On:
PubMed —
Wikipedia —
Google
Download:
MOL2
SDF
SMILES
Flexibase
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.54 |
9.17 |
-7.15 |
2 |
3 |
0 |
50 |
386.576 |
6 |
↓
|
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
Reactome Annotations from Targets (via Uniprot)
| Description |
Species |
|
Class A/1 (Rhodopsin-like receptors) |
|
|
G alpha (i) signalling events |
|
Rings
-
Cyclobutane
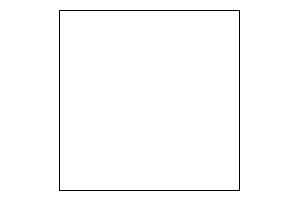
-
3,4-dihydro-2H-pyran
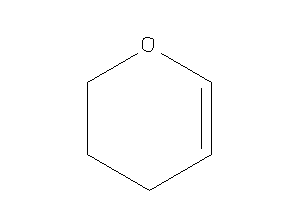
-
Benzene
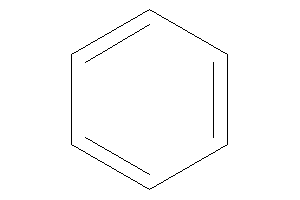
-
Cyclohexane
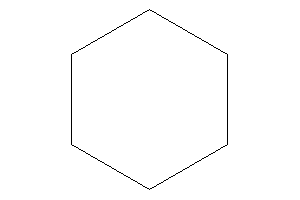
-
6a,7,8,9,10,10a-hexahydro-6H-ben…
![6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromene](//zinc12.docking.org/img/rings/188615.gif)
-
3-cyclobutyl-6a,7,8,9,10,10a-hex…
![3-cyclobutyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromene](//zinc12.docking.org/img/rings/366226.gif)
No pre-computed analogs available. Try a structural similarity search.