Substance Information
| In ZINC since |
Heavy atoms |
Benign functionality |
| January 10th, 2011 |
34 |
No
|
Popular Name:
(1S,2R,4aR,6aS,6aR,6bR,8aS,10S,11S,12aR,14bS)-10,11-dihydroxy-1,2,6a,6b,9,9,12a-heptamethyl-2,3,4,5,
(1S,2R,4aR,6aS,6aR,6bR,8aS,10S,1…
Find On:
PubMed —
Wikipedia —
Google
Download:
MOL2
SDF
SMILES
Flexibase
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.87 |
9.77 |
-53.8 |
2 |
4 |
-1 |
81 |
471.702 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.87 |
7.83 |
-7.4 |
3 |
4 |
0 |
78 |
472.71 |
1 |
↓
|
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
810 |
0.25 |
Binding ≤ 10μM
|
|
PTN1-3-E |
Protein-tyrosine Phosphatase 1B (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5490 |
0.22 |
Binding ≤ 10μM
|
|
Z80874-6-O |
CEM (T-cell Leukemia) (cluster #6 Of 7), Other |
Other |
3270 |
0.23 |
Functional ≤ 10μM |
Reactome Annotations from Targets (via Uniprot)
| Description |
Species |
|
Glucocorticoid biosynthesis |
|
|
Growth hormone receptor signaling |
|
|
Integrin alphaIIb beta3 signaling |
|
|
Regulation of IFNA signaling |
|
|
Regulation of IFNG signaling |
|
Rings
-
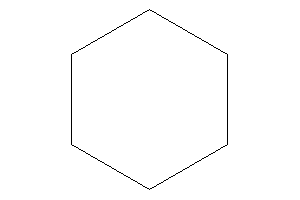
Cyclohexane
-
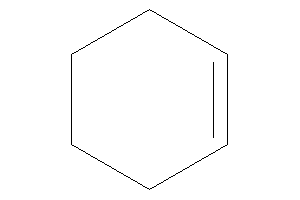
Cyclohexene
-
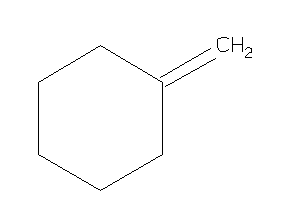
Methylenecyclohexane
-
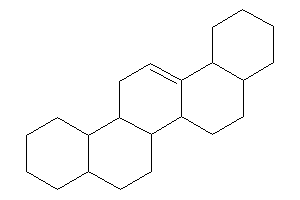
1,2,3,4,4a,5,6,6a,6a,6b,7,8,8a,9…
No pre-computed analogs available. Try a structural similarity search.