|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.60 |
Binding ≤ 10μM
|
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.60 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
410 |
0.60 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
410 |
0.60 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.60 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
414 |
0.60 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
414 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.94 |
5.37 |
-28.72 |
2 |
5 |
0 |
85 |
226.619 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.94 |
4.83 |
-35.39 |
1 |
5 |
-1 |
81 |
225.611 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-2-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
360 |
0.50 |
Binding ≤ 10μM
|
|
GRIA2-2-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
360 |
0.50 |
Binding ≤ 10μM
|
|
GRIA3-2-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
360 |
0.50 |
Binding ≤ 10μM
|
|
GRIA4-2-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
360 |
0.50 |
Binding ≤ 10μM
|
|
GRIK1-2-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.43 |
Binding ≤ 10μM |
|
GRIK2-1-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.43 |
Binding ≤ 10μM |
|
GRIK3-1-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.43 |
Binding ≤ 10μM |
|
GRIK4-1-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2900 |
0.43 |
Binding ≤ 10μM |
|
GRIK5-1-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
740 |
0.48 |
Binding ≤ 10μM
|
|
NMD3A-4-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.50 |
Binding ≤ 10μM
|
|
NMD3B-4-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.50 |
Binding ≤ 10μM
|
|
NMDE1-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
840 |
0.47 |
Binding ≤ 10μM |
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
840 |
0.47 |
Binding ≤ 10μM |
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
840 |
0.47 |
Binding ≤ 10μM |
|
NMDE4-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.50 |
Binding ≤ 10μM
|
|
NMDZ1-1-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
840 |
0.47 |
Binding ≤ 10μM |
|
Z104302-5-O |
Glutamate NMDA Receptor (cluster #5 Of 7), Other |
Other |
4500 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.46 |
1.23 |
-35.53 |
1 |
10 |
-1 |
160 |
251.134 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
0.42 |
3.67 |
-137.39 |
2 |
10 |
2 |
154 |
252.142 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.42 |
2.62 |
-51.15 |
1 |
10 |
1 |
153 |
251.134 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AIR5-1-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
72 |
0.62 |
Binding ≤ 10μM
|
|
NMD3A-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
600 |
0.54 |
Binding ≤ 10μM |
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
72 |
0.62 |
Binding ≤ 10μM
|
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.54 |
Binding ≤ 10μM |
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.54 |
Binding ≤ 10μM |
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.54 |
Binding ≤ 10μM |
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
131 |
0.60 |
Binding ≤ 10μM
|
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
600 |
0.54 |
Binding ≤ 10μM |
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
837 |
0.53 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 3), Other |
Other |
80 |
0.62 |
Functional ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 12), Other |
Other |
800 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.22 |
-0.53 |
-63.96 |
3 |
7 |
-1 |
120 |
251.199 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.22 |
1.55 |
-134.72 |
2 |
7 |
-2 |
120 |
250.191 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.22 |
0.59 |
-145.65 |
2 |
7 |
-2 |
123 |
250.191 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AIR5-1-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
72 |
0.62 |
Binding ≤ 10μM
|
|
NMD3A-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
86 |
0.62 |
Binding ≤ 10μM |
|
NMD3A-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
600 |
0.54 |
Binding ≤ 10μM |
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
72 |
0.62 |
Binding ≤ 10μM
|
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
86 |
0.62 |
Binding ≤ 10μM |
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
86 |
0.62 |
Binding ≤ 10μM |
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.54 |
Binding ≤ 10μM |
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
86 |
0.62 |
Binding ≤ 10μM |
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.54 |
Binding ≤ 10μM |
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
86 |
0.62 |
Binding ≤ 10μM |
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.54 |
Binding ≤ 10μM |
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
131 |
0.60 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3160 |
0.48 |
Binding ≤ 10μM
|
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
86 |
0.62 |
Binding ≤ 10μM |
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
600 |
0.54 |
Binding ≤ 10μM |
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1600 |
0.51 |
Functional ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1600 |
0.51 |
Functional ≤ 10μM
|
|
GRIA3-2-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.51 |
Functional ≤ 10μM
|
|
GRIA4-2-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1600 |
0.51 |
Functional ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
50 |
0.64 |
Binding ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
837 |
0.53 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 3), Other |
Other |
80 |
0.62 |
Functional ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 12), Other |
Other |
800 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.22 |
-0.58 |
-64.39 |
3 |
7 |
-1 |
120 |
251.199 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.22 |
1.56 |
-134.79 |
2 |
7 |
-2 |
120 |
250.191 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.22 |
0.54 |
-145.88 |
2 |
7 |
-2 |
123 |
250.191 |
5 |
↓
|
|
|
|
Analogs
-
26011425
-
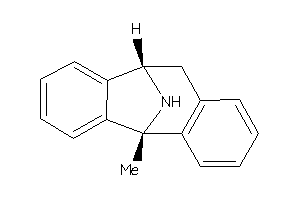
-
1482044
-
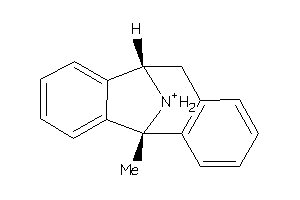
-
1854016
-
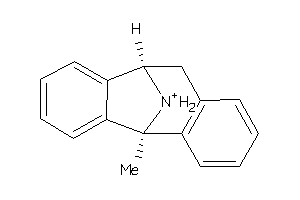
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AIR5-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.74 |
Binding ≤ 10μM
|
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.72 |
Binding ≤ 10μM
|
|
GRL1A-1-E |
Glutamate [NMDA] Receptor-like 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.72 |
Binding ≤ 10μM |
|
NMD3A-3-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.72 |
Binding ≤ 10μM |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.74 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
31 |
0.62 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
7 |
0.67 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
31 |
0.62 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7 |
0.67 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
31 |
0.62 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.67 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
31 |
0.62 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.72 |
Binding ≤ 10μM |
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
31 |
0.62 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.67 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
31 |
0.62 |
Binding ≤ 10μM
|
|
PCP-1-E |
Lysosomal Pro-X Carboxypeptidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.68 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1700 |
0.48 |
Binding ≤ 10μM
|
|
NMDE1-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.62 |
Functional ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.66 |
Functional ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
7 |
0.67 |
Binding ≤ 10μM |
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 1), Other |
Other |
5800 |
0.43 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 3), Other |
Other |
58 |
0.60 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.44 |
Functional ≤ 10μM
|
|
Z50594-6-O |
Mus Musculus (cluster #6 Of 9), Other |
Other |
6 |
0.68 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
54 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
6.6 |
-4.25 |
1 |
1 |
0 |
12 |
221.303 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE1-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
37 |
0.39 |
Binding ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
Z104298-1-O |
Glutamate NMDA Receptor; GRIN1/GRIN2A (cluster #1 Of 2), Other |
Other |
270 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.93 |
2.81 |
-46.71 |
4 |
8 |
-1 |
138 |
453.209 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.93 |
5.23 |
-81.93 |
4 |
8 |
-1 |
146 |
453.209 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.92 |
3.94 |
-132.89 |
3 |
8 |
-2 |
141 |
452.201 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE1-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
37 |
0.39 |
Binding ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
Z104298-1-O |
Glutamate NMDA Receptor; GRIN1/GRIN2A (cluster #1 Of 2), Other |
Other |
270 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.93 |
3.69 |
-44.33 |
4 |
8 |
-1 |
138 |
453.209 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.93 |
3.96 |
-38.6 |
5 |
8 |
0 |
143 |
454.217 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AIR5-1-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
730 |
0.61 |
Binding ≤ 10μM
|
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
730 |
0.61 |
Binding ≤ 10μM
|
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
912 |
0.60 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 3), Other |
Other |
800 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.74 |
0.62 |
-69.16 |
4 |
6 |
-1 |
128 |
224.173 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AIR5-1-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
NMD3A-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
461 |
0.63 |
Binding ≤ 10μM |
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
461 |
0.63 |
Binding ≤ 10μM |
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
461 |
0.63 |
Binding ≤ 10μM |
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
730 |
0.61 |
Binding ≤ 10μM
|
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
461 |
0.63 |
Binding ≤ 10μM |
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
461 |
0.63 |
Binding ≤ 10μM |
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
730 |
0.61 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
2660 |
0.56 |
Binding ≤ 10μM
|
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
461 |
0.63 |
Binding ≤ 10μM |
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
912 |
0.60 |
Binding ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
912 |
0.60 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 3), Other |
Other |
800 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.74 |
0.63 |
-68.99 |
4 |
6 |
-1 |
128 |
224.173 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3700 |
0.63 |
Binding ≤ 10μM |
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3700 |
0.63 |
Binding ≤ 10μM |
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
290 |
0.76 |
Binding ≤ 10μM
|
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3700 |
0.63 |
Binding ≤ 10μM |
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3700 |
0.63 |
Binding ≤ 10μM |
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3700 |
0.63 |
Binding ≤ 10μM |
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
176 |
0.79 |
Binding ≤ 10μM
|
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
3700 |
0.63 |
Binding ≤ 10μM |
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
350 |
0.75 |
Binding ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
420 |
0.74 |
Binding ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 12), Other |
Other |
6100 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.75 |
-0.95 |
-61.4 |
4 |
6 |
-1 |
128 |
196.119 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3700 |
0.63 |
Binding ≤ 10μM |
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3700 |
0.63 |
Binding ≤ 10μM |
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3700 |
0.63 |
Binding ≤ 10μM |
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
460 |
0.74 |
Binding ≤ 10μM
|
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3700 |
0.63 |
Binding ≤ 10μM |
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3700 |
0.63 |
Binding ≤ 10μM |
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
176 |
0.79 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3710 |
0.63 |
Binding ≤ 10μM
|
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
460 |
0.74 |
Binding ≤ 10μM
|
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
3700 |
0.63 |
Binding ≤ 10μM |
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
420 |
0.74 |
Binding ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
880 |
0.71 |
Binding ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 12), Other |
Other |
6100 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.75 |
-0.93 |
-61.42 |
4 |
6 |
-1 |
128 |
196.119 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1880 |
0.33 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1880 |
0.33 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1880 |
0.33 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1880 |
0.33 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.48 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
82 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.72 |
0.54 |
-42.47 |
2 |
2 |
1 |
24 |
322.472 |
3 |
↓
|
|
|
|
Analogs
-
1687741
-
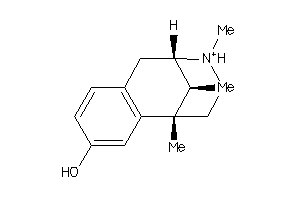
-
3954257
-
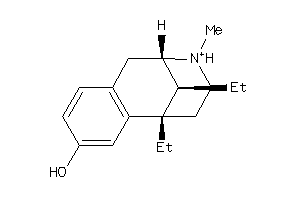
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
5960 |
0.28 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5960 |
0.28 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5960 |
0.28 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
5960 |
0.28 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.51 |
11.63 |
-41.97 |
2 |
2 |
1 |
25 |
350.526 |
5 |
↓
|
|
|
|
Analogs
-
2555353
-
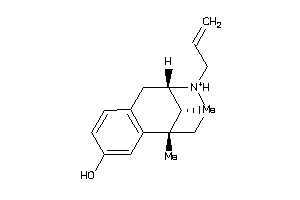
-
2555354
-
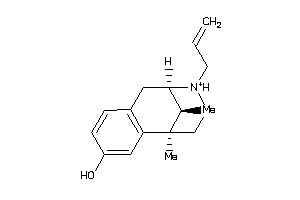
-
7996680
-
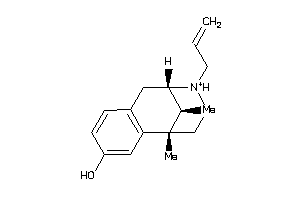
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
504 |
0.46 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.61 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.62 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
3200 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
6.87 |
-36.86 |
2 |
2 |
1 |
25 |
258.385 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2alpha,6alpha,11R*)-1,2,3,4,5,6-Hexahydro-6,11-dimethyl-2,6-methano-3-benzazocin-8-ol; 2,6-Methano-3-benzazocin-8-ol, 1,2,3,4,5,6-hexahydro-6,11-alpha-dimethyl-, hydrochloride; 2,6-Methano-3-benzazocin-8-ol, 1,2,3,4,5,6-hexahydro-6,11-dimethyl-, (2-alpha
(2alpha,6alpha,11R*)-1,2,3,4,5,6…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
30 |
0.66 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
30 |
0.66 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.66 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
30 |
0.66 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
470 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
-2.12 |
-43.7 |
3 |
2 |
1 |
36 |
218.32 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
5730 |
0.26 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3230 |
0.27 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
96 |
0.35 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1300 |
0.29 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
96 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.55 |
6.69 |
-51.34 |
2 |
7 |
-1 |
110 |
377.376 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.55 |
8.74 |
-135.8 |
1 |
7 |
-2 |
113 |
376.368 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.55 |
7.93 |
-40.92 |
3 |
7 |
0 |
114 |
378.384 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AIR5-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
95 |
0.70 |
Binding ≤ 10μM
|
|
AA3R-6-E |
Adenosine Receptor A3 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
967 |
0.60 |
Binding ≤ 10μM
|
|
GRINA-1-E |
Glutamate [NMDA] Receptor Associated Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
29 |
0.75 |
Binding ≤ 10μM |
|
NMD3A-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
69 |
0.72 |
Binding ≤ 10μM |
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
95 |
0.70 |
Binding ≤ 10μM
|
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
54 |
0.73 |
Binding ≤ 10μM |
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
54 |
0.73 |
Binding ≤ 10μM |
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
54 |
0.73 |
Binding ≤ 10μM |
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
99 |
0.70 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
54 |
0.73 |
Binding ≤ 10μM |
|
A2AIR5-1-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.58 |
Functional ≤ 10μM
|
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.58 |
Functional ≤ 10μM
|
|
NMDE1-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.58 |
Functional ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.58 |
Functional ≤ 10μM
|
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.58 |
Functional ≤ 10μM
|
|
NMDE4-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.58 |
Functional ≤ 10μM
|
|
NMDZ1-1-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1600 |
0.58 |
Functional ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
87 |
0.71 |
Binding ≤ 10μM |
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 3), Other |
Other |
22 |
0.77 |
Functional ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 12), Other |
Other |
1100 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.61 |
0.05 |
-70.08 |
3 |
6 |
-1 |
117 |
222.157 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
2150 |
0.28 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
9080 |
0.25 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4880 |
0.27 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
6030 |
0.26 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
9080 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.55 |
7.34 |
-58.43 |
2 |
7 |
-1 |
110 |
377.376 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.55 |
9.67 |
-151.96 |
1 |
7 |
-2 |
113 |
376.368 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.55 |
8.57 |
-54.76 |
3 |
7 |
0 |
114 |
378.384 |
3 |
↓
|
|
|
|
Analogs
-
1699600
-
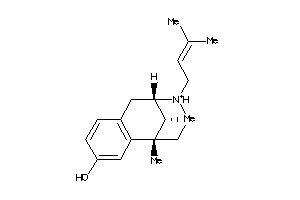
-
2015833
-
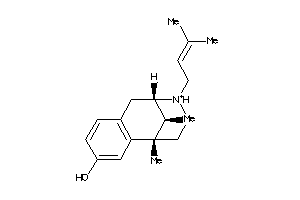
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.49 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
87 |
0.47 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.57 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
75 |
0.47 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.54 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.47 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9 |
0.54 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
87 |
0.47 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
29 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
0.88 |
-38.19 |
2 |
2 |
1 |
24 |
286.439 |
2 |
↓
|
|
|
|
Analogs
-
7996837
-
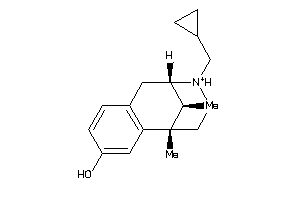
-
1227
-
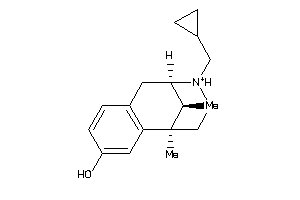
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
107 |
0.49 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
107 |
0.49 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
107 |
0.49 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
107 |
0.49 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.79 |
7.89 |
-38.06 |
2 |
2 |
1 |
25 |
272.412 |
2 |
↓
|
|
|
|
Analogs
-
596
-
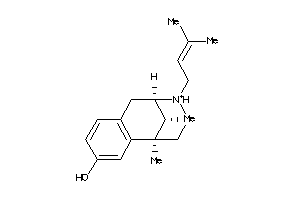
Draw
Identity
99%
90%
80%
70%
Popular Name:
(+-)-Pentazocine hydrochloride; (2R*,6R*,11R*)-1,2,3,4,5,6-Hexahydro-6,11-dimethyl-3-(3-methyl-2-butenyl)-2,6-methano-3-benzazocin-8-ol hydrochloride; (2alpha,6alpha,11R*)-1,2,3,4,5,6-Hexahydro-6,11-dimethyl-3-(3-methylbut-2-enyl)-2,6-methano-3-benzazocin
(+-)-Pentazocine hydrochloride; …
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
225 |
0.44 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
525 |
0.42 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
4320 |
0.36 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
4320 |
0.36 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4320 |
0.36 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
4320 |
0.36 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
651 |
0.41 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.56 |
Binding ≤ 10μM |
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.55 |
Binding ≤ 10μM |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
651 |
0.41 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
860 |
0.40 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
4190 |
0.36 |
Binding ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
87 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
0.88 |
-37.21 |
2 |
2 |
1 |
24 |
286.439 |
2 |
↓
|
|
|
|
Analogs
-
523
-
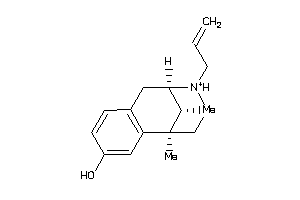
-
2555353
-

-
2555354
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(+)-SKF 10.047; (2S-(2alpha,6alpha,11R*))-1,2,3,4,5,6-Hexahydro-6,11-dimethyl-3-(2-propenyl)-2,6-methano-3-benzazocin-8-ol; 2,6-Methano-3-benzazocin-8-ol, 1,2,3,4,5,6-hexahydro-6,11-dimethyl-3-(2-propenyl)-, (2S-(2alpha,6alpha,11R*))-; C17H23NO.C2H2O4; LS
(+)-SKF 10.047; (2S-(2alpha,6alp…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
225 |
0.49 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
225 |
0.49 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
225 |
0.49 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
225 |
0.49 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
60 |
0.53 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
157 |
0.50 |
Binding ≤ 10μM |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
300 |
0.48 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
6.91 |
-38.18 |
2 |
2 |
1 |
25 |
258.385 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 120 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MURI-2-B |
Glutamate Racemase (cluster #2 Of 2), Bacterial |
Bacteria |
5800 |
0.73 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1362 |
0.82 |
Binding ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
940 |
0.84 |
Binding ≤ 10μM
|
|
GRIA3-3-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.88 |
Binding ≤ 10μM
|
|
GRIA4-3-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
868 |
0.85 |
Binding ≤ 10μM
|
|
GRIK1-3-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
701 |
0.86 |
Binding ≤ 10μM
|
|
GRIK2-2-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1106 |
0.83 |
Binding ≤ 10μM
|
|
GRIK3-2-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
789 |
0.85 |
Binding ≤ 10μM
|
|
GRIK4-2-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.90 |
Binding ≤ 10μM
|
|
GRIK5-2-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.90 |
Binding ≤ 10μM
|
|
GRM1-3-E |
Metabotropic Glutamate Receptor 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.92 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.80 |
Binding ≤ 10μM
|
|
GRM4-3-E |
Metabotropic Glutamate Receptor 4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
9800 |
0.70 |
Binding ≤ 10μM
|
|
GRM5-4-E |
Metabotropic Glutamate Receptor 5 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3100 |
0.77 |
Binding ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Binding ≤ 10μM
|
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA3-2-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA4-2-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRM1-4-E |
Metabotropic Glutamate Receptor 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1050 |
0.84 |
Functional ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.71 |
Functional ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
80 |
0.99 |
Binding ≤ 10μM |
|
Z50597-7-O |
Rattus Norvegicus (cluster #7 Of 12), Other |
Other |
2500 |
0.78 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.25 |
1.72 |
-74 |
3 |
5 |
-1 |
108 |
146.122 |
4 |
↓
|
|
|
|
Analogs
-
13726150
-
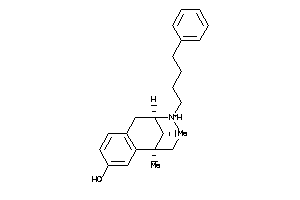
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
41 |
0.61 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
41 |
0.61 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
41 |
0.61 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
41 |
0.61 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4970 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
5.7 |
-39.28 |
2 |
2 |
1 |
25 |
232.347 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-2-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.24 |
Functional ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.23 |
Functional ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.24 |
Functional ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8000 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
12.98 |
-53.97 |
0 |
8 |
-1 |
121 |
412.381 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
34855287
-
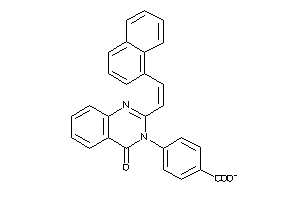
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.23 |
Functional ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.70 |
1.71 |
-54.58 |
0 |
5 |
-1 |
75 |
417.444 |
4 |
↓
|
|
|
|
Analogs
-
34720550
-
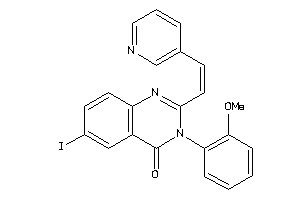
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.28 |
Functional ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.28 |
Functional ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
0.54 |
-13.72 |
0 |
5 |
0 |
57 |
481.293 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.67 |
0.64 |
-48.08 |
1 |
5 |
1 |
58 |
482.301 |
4 |
↓
|
|
|
|
Analogs
-
13087076
-
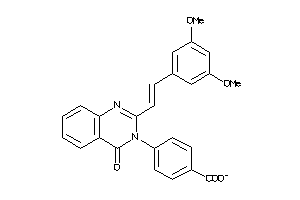
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.24 |
Functional ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.24 |
Functional ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
9000 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.75 |
1.61 |
-53.55 |
0 |
6 |
-1 |
84 |
397.41 |
5 |
↓
|
|
|
|
Analogs
-
33458634
-
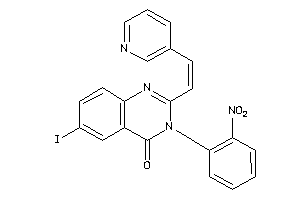
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.26 |
Functional ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.58 |
1.26 |
-18.96 |
0 |
7 |
0 |
93 |
496.264 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.58 |
1.36 |
-54.73 |
1 |
7 |
1 |
94 |
497.272 |
4 |
↓
|
|
|
|
Analogs
-
5068115
-
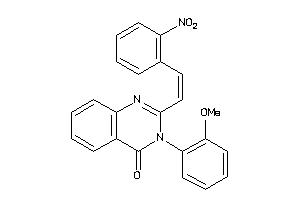
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Functional ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.24 |
Functional ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Functional ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.25 |
Functional ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.76 |
2.62 |
-20.85 |
0 |
7 |
0 |
89 |
399.406 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.22 |
Functional ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.23 |
Functional ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.69 |
13.44 |
-52.81 |
0 |
9 |
-1 |
130 |
442.407 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.25 |
Functional ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.26 |
Functional ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.74 |
15.23 |
-54.12 |
0 |
8 |
-1 |
121 |
538.277 |
5 |
↓
|
|
|
|
Analogs
-
33247734
-
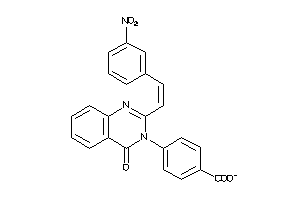
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.23 |
Functional ≤ 10μM
|
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
9000 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.66 |
14.13 |
-51.16 |
0 |
8 |
-1 |
121 |
412.381 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.34 |
5.37 |
-39.81 |
1 |
4 |
-1 |
73 |
222.607 |
1 |
↓
|
|
|
|
Analogs
-
34354758
-
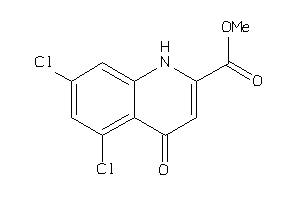
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLRA1-1-E |
Glycine Receptor Subunit Alpha-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.62 |
Binding ≤ 10μM
|
|
GLRA2-1-E |
Glycine Receptor Subunit Alpha-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.62 |
Binding ≤ 10μM
|
|
GLRA3-1-E |
Glycine Receptor Alpha-3 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.62 |
Binding ≤ 10μM
|
|
GLRB-1-E |
Glycine Receptor Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.62 |
Binding ≤ 10μM
|
|
GRIA1-2-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM |
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM |
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM |
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM |
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDE1-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDE2-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMD3B-1-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.48 |
Functional ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.48 |
Functional ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.48 |
Functional ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.48 |
Functional ≤ 10μM
|
|
Z104302-4-O |
Glutamate NMDA Receptor (cluster #4 Of 7), Other |
Other |
90 |
0.62 |
Binding ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
5000 |
0.46 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.79 |
5.87 |
-38.83 |
1 |
4 |
-1 |
73 |
257.052 |
1 |
↓
|
|