|
|
Analogs
-
120294
-
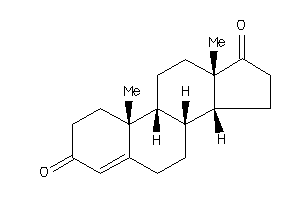
-
2046798
-
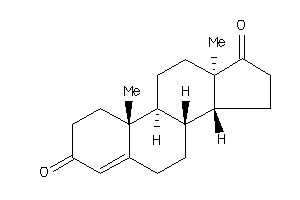
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
37 |
0.45 |
Binding ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
FABPL-1-E |
Fatty Acid-binding Protein, Liver (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.46 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
31 |
0.46 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.48 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.34 |
Binding ≤ 10μM
|
|
SGMR1-2-E |
Sigma Opioid Receptor (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
260 |
0.40 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
37 |
0.45 |
Functional ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.30 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.30 |
Functional ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2770 |
0.34 |
Functional ≤ 10μM
|
|
MCR-2-E |
Mineralocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Functional ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Functional ≤ 10μM
|
|
CP2C9-2-E |
Cytochrome P450 2C9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5500 |
0.32 |
ADME/T ≤ 10μM
|
|
ERG2-1-F |
C-8 Sterol Isomerase (cluster #1 Of 2), Fungal |
Fungi |
4430 |
0.33 |
Binding ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
6310 |
0.32 |
Functional ≤ 10μM
|
|
Z80712-1-O |
T47D (Breast Carcinoma Cells) (cluster #1 Of 7), Other |
Other |
1 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
10.64 |
-9.9 |
0 |
2 |
0 |
34 |
314.469 |
1 |
↓
|
|
|
|
Analogs
-
4257980
-
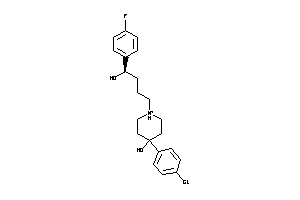
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
8.57 |
-46.63 |
3 |
3 |
1 |
45 |
378.895 |
6 |
↓
|
|
|
|
Analogs
-
2516030
-
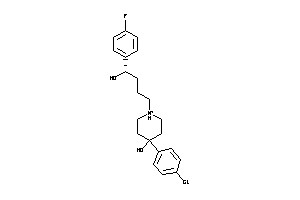
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
8.56 |
-46.59 |
3 |
3 |
1 |
45 |
378.895 |
6 |
↓
|
|
|
|
Analogs
-
14551
-
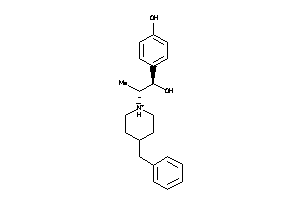
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.41 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.48 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
100 |
0.41 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
21 |
0.45 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
73 |
0.42 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.48 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.41 |
Functional ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
110 |
0.41 |
Functional ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
1 |
0.53 |
Binding ≤ 10μM
|
|
Z104297-2-O |
Glutamate NMDA Receptor; GRIN1/GRIN2B (cluster #2 Of 3), Other |
Other |
110 |
0.41 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 3), Other |
Other |
470 |
0.37 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.32 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
26 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
7.84 |
-40 |
3 |
3 |
1 |
45 |
326.46 |
5 |
↓
|
|
|
|
Analogs
-
36779566
-
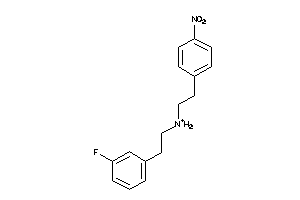
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2254 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.39 |
0.75 |
-54.34 |
2 |
4 |
1 |
62 |
209.269 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.36 |
Binding ≤ 10μM |
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
352 |
0.32 |
Binding ≤ 10μM |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
331 |
0.32 |
Binding ≤ 10μM |
|
DRD2-4-E |
Dopamine D2 Receptor (cluster #4 Of 24), Eukaryotic |
Eukaryotes |
71 |
0.36 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.02 |
-1.97 |
-38.24 |
1 |
4 |
1 |
27 |
392.548 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.41 |
Binding ≤ 10μM |
|
DRD4-2-E |
Dopamine D4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
652 |
0.36 |
Binding ≤ 10μM |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.46 |
Binding ≤ 10μM |
|
DRD2-23-E |
Dopamine D2 Receptor (cluster #23 Of 24), Eukaryotic |
Eukaryotes |
5 |
0.48 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
9.17 |
-43.44 |
3 |
3 |
1 |
40 |
386.313 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.12 |
6.89 |
-8.82 |
2 |
3 |
0 |
39 |
385.305 |
3 |
↓
|
|
|
|
Analogs
-
39318902
-
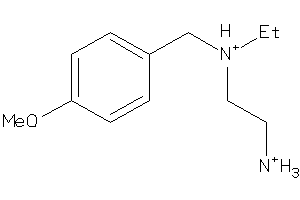
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
161 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
8.7 |
-38.9 |
1 |
3 |
1 |
17 |
297.422 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.09 |
8.64 |
-39.12 |
1 |
3 |
1 |
17 |
297.422 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.09 |
6.32 |
-4.57 |
0 |
3 |
0 |
16 |
296.414 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.45 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
11.07 |
-45.98 |
1 |
4 |
1 |
26 |
369.529 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.01 |
8.76 |
-7.53 |
0 |
4 |
0 |
25 |
368.521 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.01 |
11.13 |
-44.47 |
1 |
4 |
1 |
26 |
369.529 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.35 |
3.41 |
-41.2 |
1 |
3 |
1 |
22 |
356.53 |
12 |
↓
|
|
|
|
Analogs
-
359759
-
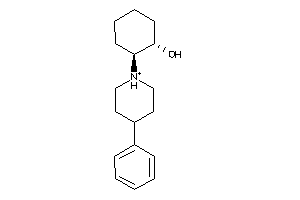
-
359762
-
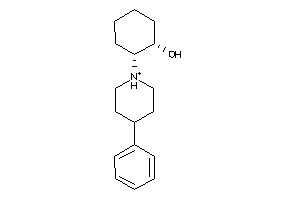
-
3173766
-
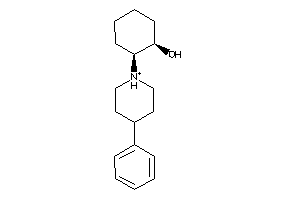
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
34 |
0.55 |
Binding ≤ 10μM
|
|
VACHT-1-E |
Vesicular Acetylcholine Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.62 |
Binding ≤ 10μM |
|
VACHT-1-E |
Vesicular Acetylcholine Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
34 |
0.55 |
Binding ≤ 10μM
|
|
VACHT-1-E |
Vesicular Acetylcholine Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.55 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
34 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
7.72 |
-33.08 |
2 |
2 |
1 |
25 |
260.401 |
2 |
↓
|
|
|
|
Analogs
-
8143662
-
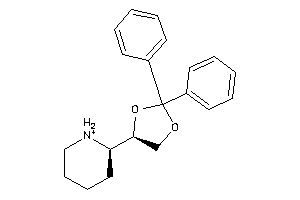
-
8659933
-
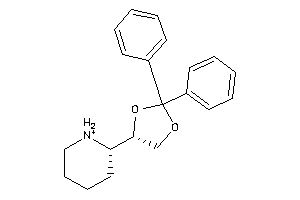
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-3-E |
Sigma Opioid Receptor (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
35 |
0.45 |
Binding ≤ 10μM
|
|
Z104302-2-O |
Glutamate NMDA Receptor (cluster #2 Of 7), Other |
Other |
104 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
9.95 |
-37.44 |
2 |
3 |
1 |
35 |
310.417 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.04 |
8.99 |
-3.84 |
1 |
3 |
0 |
30 |
309.409 |
3 |
↓
|
|
|
|
Analogs
-
8143662
-

-
8659933
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-3-E |
Sigma Opioid Receptor (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
35 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
9.97 |
-36.73 |
2 |
3 |
1 |
35 |
310.417 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.04 |
8.69 |
-4.54 |
1 |
3 |
0 |
30 |
309.409 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.40 |
11.82 |
-38.46 |
1 |
2 |
1 |
8 |
309.477 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.40 |
11.84 |
-36.2 |
1 |
2 |
1 |
8 |
309.477 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.40 |
9.52 |
-3.62 |
0 |
2 |
0 |
6 |
308.469 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.72 |
11.82 |
-38.09 |
1 |
2 |
1 |
8 |
374.346 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.72 |
11.76 |
-31.4 |
1 |
2 |
1 |
8 |
374.346 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.72 |
9.5 |
-3.7 |
0 |
2 |
0 |
6 |
373.338 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.09 |
11.03 |
-39.53 |
1 |
2 |
1 |
8 |
313.44 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.09 |
11.1 |
-41.2 |
1 |
2 |
1 |
8 |
313.44 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.09 |
8.72 |
-4.37 |
0 |
2 |
0 |
6 |
312.432 |
6 |
↓
|
|
|
|
Analogs
-
37010517
-
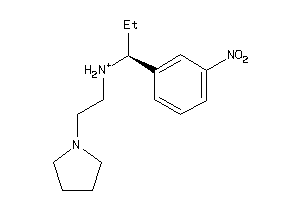
-
37010518
-
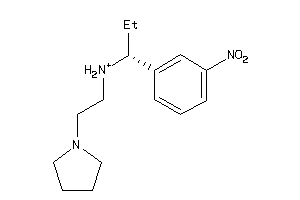
-
4703393
-
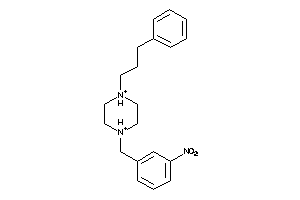
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
11.83 |
-45.9 |
1 |
5 |
1 |
53 |
340.447 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.89 |
11.83 |
-51.11 |
1 |
5 |
1 |
53 |
340.447 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.89 |
9.51 |
-8.72 |
0 |
5 |
0 |
52 |
339.439 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
10.47 |
-40.5 |
1 |
3 |
1 |
17 |
325.476 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.01 |
10.48 |
-39.32 |
1 |
3 |
1 |
17 |
325.476 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.01 |
8.16 |
-5.01 |
0 |
3 |
0 |
16 |
324.468 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.87 |
12.51 |
-44.7 |
1 |
5 |
1 |
53 |
340.447 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.87 |
11.81 |
-35.77 |
1 |
5 |
1 |
53 |
340.447 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.87 |
10.19 |
-9.2 |
0 |
5 |
0 |
52 |
339.439 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
10.84 |
-37.93 |
1 |
2 |
1 |
8 |
295.45 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.96 |
10.86 |
-36.22 |
1 |
2 |
1 |
8 |
295.45 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.96 |
8.47 |
-3.29 |
0 |
2 |
0 |
6 |
294.442 |
6 |
↓
|
|
|
|
Analogs
-
45650983
-
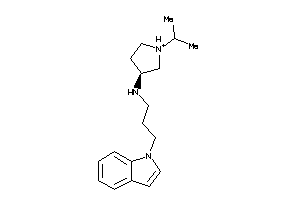
-
45650984
-
Draw
Identity
99%
90%
80%
70%
Vendors
-
-
- API0009158
- CHM0162901
- CHM0145732
-
And 10 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
9.85 |
-38.47 |
2 |
3 |
1 |
25 |
322.476 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.14 |
12.13 |
-120.85 |
3 |
3 |
2 |
26 |
323.484 |
4 |
↓
|
|
|
|
Analogs
-
40164019
-
-
40164021
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
744 |
0.48 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2960 |
0.43 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7660 |
0.40 |
Binding ≤ 10μM
|
|
NMD3A-6-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMD3B-6-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE1-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4-6-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDZ1-6-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
OPRD-3-E |
Delta Opioid Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3950 |
0.42 |
Binding ≤ 10μM
|
|
OPRK-2-E |
Kappa Opioid Receptor (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3950 |
0.42 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3950 |
0.42 |
Binding ≤ 10μM
|
|
SGMR1-4-E |
Sigma Opioid Receptor (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
89 |
0.55 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9210 |
0.39 |
Functional ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
2000 |
0.44 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
70 |
0.56 |
Binding ≤ 10μM
|
|
Z104302-7-O |
Glutamate NMDA Receptor (cluster #7 Of 7), Other |
Other |
9120 |
0.39 |
Binding ≤ 10μM
|
|
Z50512-3-O |
Cavia Porcellus (cluster #3 Of 7), Other |
Other |
6900 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
8.4 |
-25.53 |
4 |
3 |
1 |
50 |
240.33 |
4 |
↓
|
|
|
|
Analogs
-
14551
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
7.74 |
-38.94 |
3 |
3 |
1 |
45 |
326.46 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.48 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.17 |
9.85 |
-2.91 |
0 |
2 |
0 |
12 |
303.49 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
10.48 |
-39.39 |
1 |
3 |
1 |
17 |
325.476 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.99 |
10.48 |
-37.79 |
1 |
3 |
1 |
17 |
325.476 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.99 |
8.15 |
-5.25 |
0 |
3 |
0 |
16 |
324.468 |
7 |
↓
|
|
|
|
Analogs
-
2734776
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.91 |
11.82 |
-48.57 |
1 |
5 |
1 |
53 |
340.447 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.91 |
11.83 |
-53.24 |
1 |
5 |
1 |
53 |
340.447 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.91 |
9.51 |
-7.82 |
0 |
5 |
0 |
52 |
339.439 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
29 |
0.39 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.32 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.45 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
14 |
0.41 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
71 |
0.37 |
Binding ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
604 |
0.32 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
9 |
0.42 |
Binding ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
1750 |
0.30 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
2620 |
0.29 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
38 |
0.38 |
Functional ≤ 10μM
|
|
Z80492-2-O |
SK-N-SH (Neuroblastoma Cells) (cluster #2 Of 2), Other |
Other |
9370 |
0.26 |
Functional ≤ 10μM
|
|
Z80492-2-O |
SK-N-SH (Neuroblastoma Cells) (cluster #2 Of 2), Other |
Other |
9970 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.07 |
12.01 |
-35.57 |
1 |
3 |
1 |
17 |
371.589 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.07 |
9.8 |
-3.91 |
0 |
3 |
0 |
16 |
370.581 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6300 |
0.30 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
695 |
0.36 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6570 |
0.30 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
722 |
0.36 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
87 |
0.41 |
Binding ≤ 10μM
|
|
DRD4-2-E |
Dopamine D4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
449 |
0.37 |
Binding ≤ 10μM |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
19 |
0.45 |
Binding ≤ 10μM |
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Functional ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
90 |
0.41 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.32 |
Functional ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
9.29 |
-42.24 |
3 |
3 |
1 |
40 |
341.862 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.99 |
7.06 |
-6.67 |
2 |
3 |
0 |
39 |
340.854 |
3 |
↓
|
|
|
|
Analogs
-
34826692
-
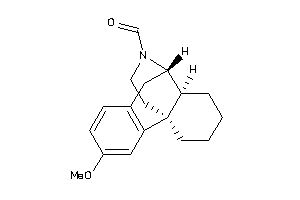
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-3-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMD3A-3-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
490 |
0.44 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1680 |
0.40 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5070 |
0.37 |
Binding ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3400 |
0.38 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5600 |
0.37 |
ADME/T ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
590 |
0.44 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
2246 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
794 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
8.86 |
-39.89 |
1 |
2 |
1 |
14 |
272.412 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
29 |
0.39 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.32 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6 |
0.43 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
14 |
0.41 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
71 |
0.37 |
Binding ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
604 |
0.32 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
2 |
0.45 |
Binding ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
1500 |
0.30 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
2620 |
0.29 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
38 |
0.38 |
Functional ≤ 10μM
|
|
Z80492-2-O |
SK-N-SH (Neuroblastoma Cells) (cluster #2 Of 2), Other |
Other |
9070 |
0.26 |
Functional ≤ 10μM
|
|
Z80492-2-O |
SK-N-SH (Neuroblastoma Cells) (cluster #2 Of 2), Other |
Other |
9970 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.07 |
12.02 |
-35.51 |
1 |
3 |
1 |
17 |
371.589 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.07 |
9.8 |
-3.9 |
0 |
3 |
0 |
16 |
370.581 |
6 |
↓
|
|
|
|
Analogs
-
5750285
-
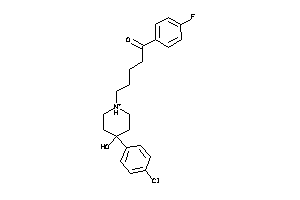
-
36464350
-
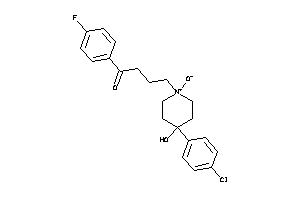
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.28 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.34 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.34 |
Binding ≤ 10μM
|
|
5HT1E-1-E |
Serotonin 1e (5-HT1e) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.34 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.34 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.41 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
27 |
0.41 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
263 |
0.35 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
5500 |
0.28 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6120 |
0.28 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4670 |
0.29 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4670 |
0.29 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4670 |
0.29 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
19 |
0.42 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9600 |
0.27 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9600 |
0.27 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9600 |
0.27 |
Binding ≤ 10μM
|
|
AP2S1-1-E |
AP-2 Complex Subunit Sigma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
24 |
0.41 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.34 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
DRD4-3-E |
Dopamine D4 Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
13 |
0.42 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.43 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
190 |
0.36 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
730 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7520 |
0.28 |
Binding ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5300 |
0.28 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5687 |
0.28 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5687 |
0.28 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5687 |
0.28 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.28 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1800 |
0.31 |
Binding ≤ 10μM
|
|
SCN1A-2-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.32 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.32 |
Binding ≤ 10μM
|
|
SCN3A-2-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.32 |
Binding ≤ 10μM
|
|
SCN8A-2-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.32 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
550 |
0.34 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
28 |
0.41 |
Functional ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
1 |
0.48 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
70 |
0.39 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
7466 |
0.28 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
9 |
0.43 |
Binding ≤ 10μM
|
|
Z103204-1-O |
A427 (cluster #1 Of 4), Other |
Other |
10000 |
0.27 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3162 |
0.30 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
7000 |
0.28 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
10 |
0.43 |
Functional ≤ 10μM
|
|
Z80091-1-O |
CHO-hD3 (cluster #1 Of 1), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Q4U254-1-V |
Human Rhinovirus A Protease (cluster #1 Of 3), Viral |
Viruses |
100 |
0.38 |
Binding ≤ 10μM
|
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
100 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
10.74 |
-49.71 |
2 |
3 |
1 |
42 |
376.879 |
6 |
↓
|
|
|
|
Analogs
-
1699600
-
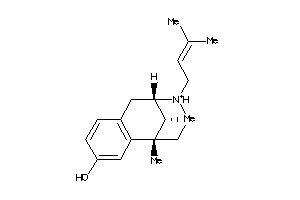
-
2015833
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.49 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
87 |
0.47 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.57 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
75 |
0.47 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.54 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.47 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9 |
0.54 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
87 |
0.47 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
29 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
0.88 |
-38.19 |
2 |
2 |
1 |
24 |
286.439 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
37038920
-
-
37038921
-
-
37038922
-
-
37038923
-
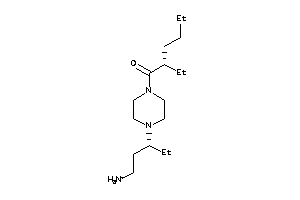
-
37038996
-
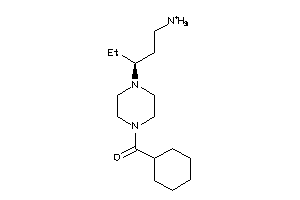
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.46 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
7.37 |
-6.49 |
0 |
3 |
0 |
24 |
278.44 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.75 |
9.6 |
-44.81 |
1 |
3 |
1 |
25 |
279.448 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
Z80492-2-O |
SK-N-SH (Neuroblastoma Cells) (cluster #2 Of 2), Other |
Other |
1400 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.63 |
15.09 |
-35.57 |
1 |
2 |
1 |
14 |
370.601 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
Z80492-2-O |
SK-N-SH (Neuroblastoma Cells) (cluster #2 Of 2), Other |
Other |
1400 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.63 |
15.08 |
-35.75 |
1 |
2 |
1 |
14 |
370.601 |
6 |
↓
|
|
|
|
Analogs
-
596
-
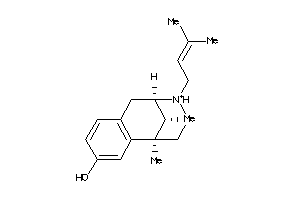
Draw
Identity
99%
90%
80%
70%
Popular Name:
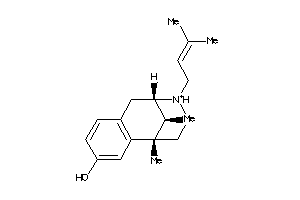
(+-)-Pentazocine hydrochloride; (2R*,6R*,11R*)-1,2,3,4,5,6-Hexahydro-6,11-dimethyl-3-(3-methyl-2-butenyl)-2,6-methano-3-benzazocin-8-ol hydrochloride; (2alpha,6alpha,11R*)-1,2,3,4,5,6-Hexahydro-6,11-dimethyl-3-(3-methylbut-2-enyl)-2,6-methano-3-benzazocin
(+-)-Pentazocine hydrochloride; …
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
225 |
0.44 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
525 |
0.42 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
4320 |
0.36 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
4320 |
0.36 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4320 |
0.36 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
4320 |
0.36 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
651 |
0.41 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.56 |
Binding ≤ 10μM |
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.55 |
Binding ≤ 10μM |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
651 |
0.41 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
860 |
0.40 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
4190 |
0.36 |
Binding ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
87 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
0.88 |
-37.21 |
2 |
2 |
1 |
24 |
286.439 |
2 |
↓
|
|
|
|
Analogs
-
3995923
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
320 |
0.36 |
Binding ≤ 10μM |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
340 |
0.36 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
410 |
0.36 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
410 |
0.36 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
410 |
0.36 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
60 |
0.40 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
490 |
0.35 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
112 |
0.39 |
Functional ≤ 10μM
|
|
DRD2-6-E |
Dopamine D2 Receptor (cluster #6 Of 24), Eukaryotic |
Eukaryotes |
3346 |
0.31 |
Binding ≤ 10μM
|
|
DRD2-6-E |
Dopamine D2 Receptor (cluster #6 Of 24), Eukaryotic |
Eukaryotes |
8400 |
0.28 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
96 |
0.39 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
230 |
0.37 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
520 |
0.35 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
570 |
0.35 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
-1.92 |
-45.78 |
2 |
5 |
1 |
53 |
349.405 |
6 |
↓
|
|
|
|
Analogs
-
13635
-
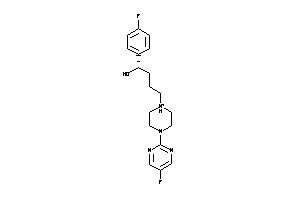
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
210 |
0.37 |
Binding ≤ 10μM
|
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
320 |
0.36 |
Binding ≤ 10μM |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
410 |
0.36 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
410 |
0.36 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
410 |
0.36 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
60 |
0.40 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
64 |
0.40 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
112 |
0.39 |
Functional ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
230 |
0.37 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
1235 |
0.33 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
520 |
0.35 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
610 |
0.35 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
DRD2-6-E |
Dopamine D2 Receptor (cluster #6 Of 24), Eukaryotic |
Eukaryotes |
8400 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
9.96 |
-46.56 |
2 |
5 |
1 |
54 |
349.405 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.69 |
7.76 |
-7.98 |
1 |
5 |
0 |
52 |
348.397 |
6 |
↓
|
|
|
|
Analogs
-
34627526
-
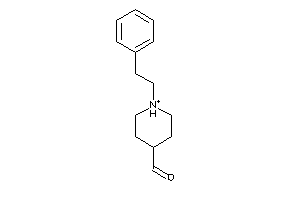
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
595 |
0.62 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
89 |
0.71 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
194 |
0.67 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
9.12 |
-34.28 |
1 |
1 |
1 |
4 |
190.31 |
3 |
↓
|
|