|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-1-B |
Beta-lactamase (cluster #1 Of 6), Bacterial |
Bacteria |
6 |
0.46 |
Binding ≤ 10μM
|
|
CP51-1-B |
Sterol 14-alpha Demethylase (cluster #1 Of 2), Bacterial |
Bacteria |
200 |
0.38 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
82 |
0.40 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
130 |
0.39 |
Binding ≤ 10μM
|
|
KCNA3-1-E |
Voltage-gated Potassium Channel Subunit Kv1.3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.29 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3020 |
0.31 |
Binding ≤ 10μM
|
|
KCNN4-1-E |
Intermediate Conductance Calcium-activated Potassium Channel Protein 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.40 |
Binding ≤ 10μM
|
|
MDHM-1-E |
Malate Dehydrogenase, Mitochondrial (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6700 |
0.29 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4800 |
0.30 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.41 |
ADME/T ≤ 10μM
|
|
Q96W81-1-F |
14-alpha Sterol Demethylase (cluster #1 Of 1), Fungal |
Fungi |
103 |
0.39 |
Binding ≤ 10μM
|
|
Q4WNT5-1-O |
14-alpha Sterol Demethylase Cyp51A (cluster #1 Of 1), Other |
Other |
4790 |
0.30 |
Binding ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
6540 |
0.29 |
Functional ≤ 10μM
|
|
Z50425-9-O |
Plasmodium Falciparum (cluster #9 Of 22), Other |
Other |
60 |
0.40 |
Functional ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
250 |
0.37 |
Functional ≤ 10μM
|
|
Z80682-8-O |
A549 (Lung Carcinoma Cells) (cluster #8 Of 11), Other |
Other |
5100 |
0.30 |
Functional ≤ 10μM
|
|
Z80951-2-O |
NIH3T3 (Fibroblasts) (cluster #2 Of 4), Other |
Other |
630 |
0.35 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
3000 |
0.31 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.47 |
13.43 |
-5.78 |
0 |
2 |
0 |
18 |
344.845 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.47 |
13.99 |
-28.18 |
1 |
2 |
1 |
19 |
345.853 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
8214681
-
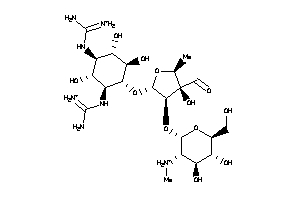
-
8214760
-
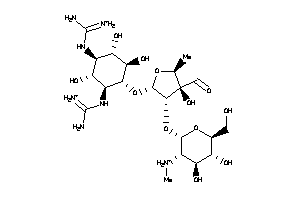
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TARB1-1-E |
TAR RNA Binding Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9500 |
0.18 |
Binding ≤ 10μM |
|
TRBP2-1-E |
TAR RNA Binding Protein 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9500 |
0.18 |
Binding ≤ 10μM |
|
Z50212-7-O |
Escherichia Coli (cluster #7 Of 7), Other |
Other |
1050 |
0.21 |
Functional ≤ 10μM
|
|
Z50380-2-O |
Mycobacterium Smegmatis (cluster #2 Of 4), Other |
Other |
500 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.28 |
-15.28 |
-142.69 |
19 |
19 |
3 |
339 |
584.604 |
11 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.28 |
-16.72 |
-90.55 |
18 |
19 |
2 |
335 |
583.596 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50380-2-O |
Mycobacterium Smegmatis (cluster #2 Of 4), Other |
Other |
2120 |
0.17 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50380-2-O |
Mycobacterium Smegmatis (cluster #2 Of 4), Other |
Other |
2120 |
0.17 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
4262458
-
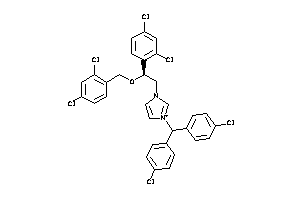
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LUXR-1-B |
Transcriptional Activator Protein LuxR (cluster #1 Of 2), Bacterial |
Bacteria |
7000 |
0.18 |
Binding ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
7020 |
0.18 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.18 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.78 |
24.12 |
-44.27 |
0 |
3 |
1 |
18 |
652.256 |
9 |
↓
|
|
|
|
Analogs
-
4262457
-
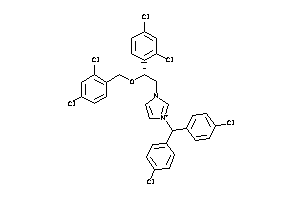
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LUXR-1-B |
Transcriptional Activator Protein LuxR (cluster #1 Of 2), Bacterial |
Bacteria |
7000 |
0.18 |
Binding ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
7020 |
0.18 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.18 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.78 |
24.09 |
-46.1 |
0 |
3 |
1 |
18 |
652.256 |
9 |
↓
|
|
|
|
|
|
|
Analogs
-
5820628
-
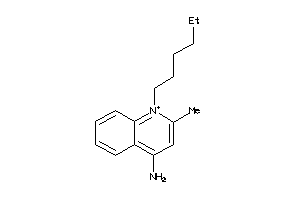
-
2583341
-
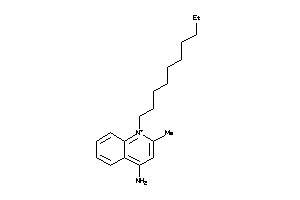
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50380-3-O |
Mycobacterium Smegmatis (cluster #3 Of 4), Other |
Other |
3950 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.13 |
18.96 |
-54.49 |
4 |
4 |
2 |
60 |
512.786 |
15 |
↓
|
|
|
|
Analogs
-
16248873
-
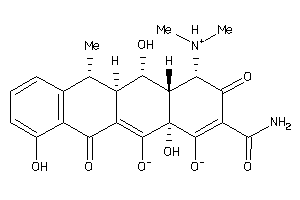
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.43 |
0.98 |
-89.55 |
6 |
10 |
-1 |
188 |
443.432 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-0.56 |
-4.34 |
-53.29 |
6 |
10 |
-1 |
185 |
443.432 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.61 |
-2.24 |
-69.99 |
6 |
10 |
-1 |
184 |
443.432 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.24 |
0.59 |
-95.46 |
6 |
10 |
-1 |
188 |
443.432 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.69 |
-1.58 |
-146.85 |
5 |
10 |
-2 |
187 |
442.424 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.24 |
-0.62 |
-247.57 |
4 |
10 |
-3 |
190 |
441.416 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
7660 |
0.65 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.46 |
3.39 |
-12.08 |
2 |
2 |
0 |
39 |
166.249 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.46 |
3.72 |
-44.66 |
3 |
2 |
1 |
40 |
167.257 |
2 |
↓
|
|
|
|
Analogs
-
34659298
-
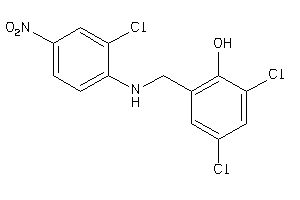
Draw
Identity
99%
90%
80%
70%
Vendors
And 42 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
JAK2-1-E |
Tyrosine-protein Kinase JAK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM |
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM |
|
STAT3-1-E |
Signal Transducer And Activator Of Transcription 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
250 |
0.44 |
Binding ≤ 10μM |
|
Z100499-1-O |
SARS Coronavirus (cluster #1 Of 2), Other |
Other |
100 |
0.47 |
Functional ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
8800 |
0.34 |
Functional ≤ 10μM |
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
6850 |
0.34 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
794 |
0.41 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
700 |
0.41 |
Functional ≤ 10μM |
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
7200 |
0.34 |
Functional ≤ 10μM |
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
700 |
0.41 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
400 |
0.43 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
400 |
0.43 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
1400 |
0.39 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.54 |
6.79 |
-7.83 |
2 |
6 |
0 |
95 |
327.123 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.54 |
7.79 |
-39.71 |
1 |
6 |
-1 |
98 |
326.115 |
3 |
↓
|
|
|
|
Analogs
-
34013998
-
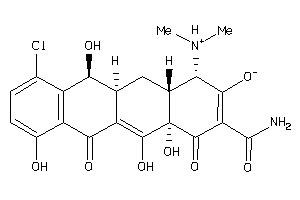
-
13719774
-
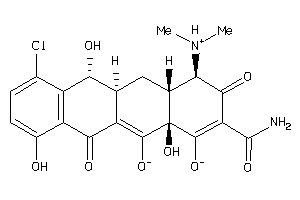
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50380-2-O |
Mycobacterium Smegmatis (cluster #2 Of 4), Other |
Other |
5920 |
0.23 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
794 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.38 |
-2.26 |
-63.05 |
6 |
10 |
-1 |
184 |
463.85 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
-0.93 |
-2.27 |
-62.34 |
6 |
10 |
-1 |
184 |
463.85 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.38 |
-1.28 |
-142.66 |
5 |
10 |
-2 |
187 |
462.842 |
1 |
↓
|
|
|
|
Analogs
-
19868839
-
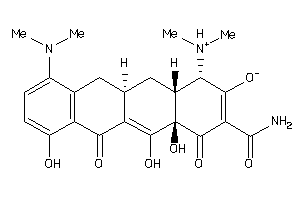
-
6566166
-
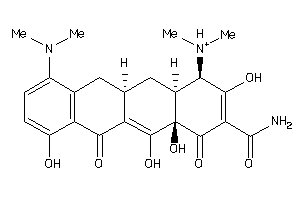
-
8691805
-
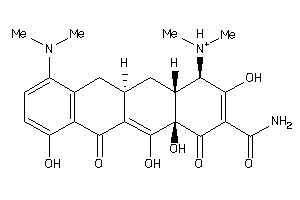
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.23 |
3.53 |
-52.8 |
6 |
10 |
0 |
169 |
457.483 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.23 |
2.49 |
-137.36 |
4 |
10 |
-2 |
170 |
455.467 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.96 |
-0.02 |
-17.13 |
6 |
10 |
0 |
165 |
457.483 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
7996793
-
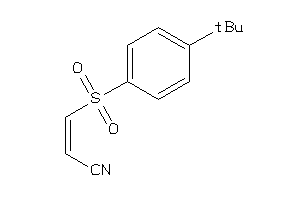
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
6220 |
0.43 |
Functional ≤ 10μM
|
|
Z50425-2-O |
Plasmodium Falciparum (cluster #2 Of 22), Other |
Other |
6310 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
5.59 |
-9.72 |
0 |
3 |
0 |
58 |
249.335 |
3 |
↓
|
|
|
|
Analogs
-
538273
-
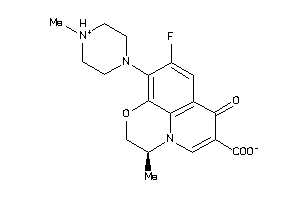
-
22056311
-
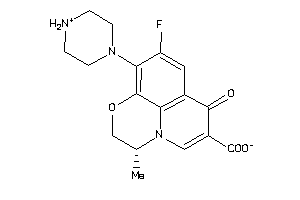
-
22056381
-
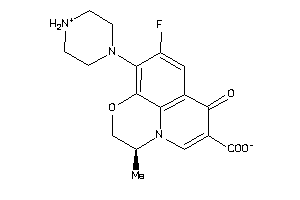
Draw
Identity
99%
90%
80%
70%
Vendors
And 51 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.26 |
10.08 |
-92.39 |
1 |
7 |
0 |
79 |
361.373 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.26 |
7.74 |
-63.35 |
0 |
7 |
-1 |
78 |
360.365 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.01 |
8.22 |
-73.63 |
2 |
7 |
1 |
82 |
362.381 |
1 |
↓
|
|
|
|
Analogs
-
22056311
-

-
22056381
-

-
537891
-
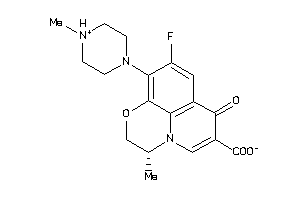
Draw
Identity
99%
90%
80%
70%
Vendors
And 69 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.26 |
9.75 |
-92.26 |
1 |
7 |
0 |
79 |
361.373 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.01 |
7.88 |
-73.44 |
2 |
7 |
1 |
82 |
362.381 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.26 |
7.41 |
-63.01 |
0 |
7 |
-1 |
78 |
360.365 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 150 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
5.39 |
-13.16 |
0 |
2 |
0 |
26 |
180.21 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.90 |
5.91 |
-29.73 |
1 |
2 |
1 |
27 |
181.218 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 37 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
411 |
0.32 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
BGLR-2-E |
Beta-glucuronidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
CALM-2-E |
Calmodulin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
LYSC1-1-E |
Lysozyme C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.25 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6500 |
0.26 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
15 |
0.39 |
Binding ≤ 10μM
|
|
DRD2-14-E |
Dopamine D2 Receptor (cluster #14 Of 24), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
500 |
0.32 |
Binding ≤ 10μM
|
|
PDR5-1-F |
Pleiotropic ABC Efflux Transporter Of Multiple Drugs (cluster #1 Of 1), Fungal |
Fungi |
1400 |
0.29 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
350 |
0.32 |
Binding ≤ 10μM
|
|
Z100748-2-O |
Neutrophils (cluster #2 Of 2), Other |
Other |
6600 |
0.26 |
Functional ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
3150 |
0.28 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6000 |
0.26 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
6600 |
0.26 |
Functional ≤ 10μM
|
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
700 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.14 |
11.43 |
-38.84 |
1 |
3 |
1 |
13 |
408.513 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.14 |
9.06 |
-5.72 |
0 |
3 |
0 |
11 |
407.505 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.14 |
11.32 |
-44.3 |
1 |
3 |
1 |
13 |
408.513 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50380-4-O |
Mycobacterium Smegmatis (cluster #4 Of 4), Other |
Other |
3660 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.35 |
-1.34 |
-108.47 |
6 |
4 |
2 |
74 |
206.33 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.23 |
18.16 |
-206.41 |
8 |
6 |
4 |
85 |
586.95 |
29 |
↓
|
|
Hi
High (pH 8-9.5)
|
7.23 |
16.77 |
-145.93 |
7 |
6 |
3 |
80 |
585.942 |
29 |
↓
|
|
|
|
Analogs
-
6535287
-
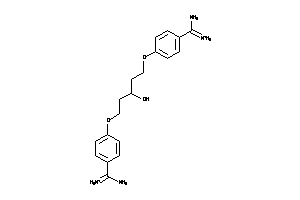
-
6535291
-
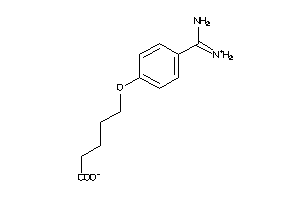
-
6535292
-
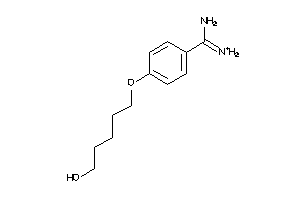
-
34928597
-
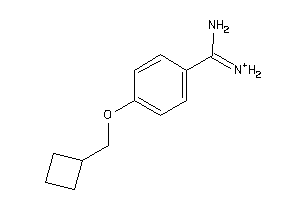
Draw
Identity
99%
90%
80%
70%
Vendors
And 50 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S100B-2-E |
S-100 Protein Beta Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.34 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1160 |
0.33 |
Binding ≤ 10μM
|
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2300 |
0.32 |
Binding ≤ 10μM
|
|
Z104302-2-O |
Glutamate NMDA Receptor (cluster #2 Of 7), Other |
Other |
3060 |
0.31 |
Binding ≤ 10μM |
|
Z101865-1-O |
Leishmania Chagasi (cluster #1 Of 1), Other |
Other |
1110 |
0.33 |
Functional ≤ 10μM
|
|
Z101870-2-O |
Leishmania Guyanensis (cluster #2 Of 2), Other |
Other |
900 |
0.34 |
Functional ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
84 |
0.40 |
Functional ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
9100 |
0.28 |
Functional ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
6 |
0.46 |
Functional ≤ 10μM
|
|
Z50420-1-O |
Trypanosoma Brucei Brucei (cluster #1 Of 7), Other |
Other |
3 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-9-O |
Plasmodium Falciparum (cluster #9 Of 22), Other |
Other |
251 |
0.37 |
Functional ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
6 |
0.46 |
Functional ≤ 10μM
|
|
Z50457-1-O |
Leishmania Amazonensis (cluster #1 Of 3), Other |
Other |
460 |
0.35 |
Functional ≤ 10μM
|
|
Z50458-4-O |
Leishmania Braziliensis (cluster #4 Of 4), Other |
Other |
9800 |
0.28 |
Functional ≤ 10μM
|
|
Z50459-1-O |
Leishmania Donovani (cluster #1 Of 8), Other |
Other |
9800 |
0.28 |
Functional ≤ 10μM
|
|
Z50460-1-O |
Leishmania Major (cluster #1 Of 4), Other |
Other |
7500 |
0.29 |
Functional ≤ 10μM
|
|
Z50461-1-O |
Leishmania Mexicana (cluster #1 Of 3), Other |
Other |
9568 |
0.28 |
Functional ≤ 10μM
|
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
7100 |
0.29 |
Functional ≤ 10μM
|
|
Z50467-2-O |
Trichomonas Vaginalis (cluster #2 Of 3), Other |
Other |
3815 |
0.30 |
Functional ≤ 10μM
|
|
Z50468-4-O |
Giardia Intestinalis (cluster #4 Of 4), Other |
Other |
4079 |
0.30 |
Functional ≤ 10μM
|
|
Z50473-4-O |
Plasmodium Berghei (cluster #4 Of 5), Other |
Other |
2942 |
0.31 |
Functional ≤ 10μM
|
|
Z50594-6-O |
Mus Musculus (cluster #6 Of 9), Other |
Other |
3 |
0.48 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
2500 |
0.31 |
Functional ≤ 10μM
|
|
Z50725-1-O |
Trypanosoma Brucei Rhodesiense (cluster #1 Of 7), Other |
Other |
50 |
0.41 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
40 |
0.41 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
2100 |
0.32 |
Functional ≤ 10μM
|
|
Z80178-2-O |
J774.A1 (Macrophage Cells) (cluster #2 Of 2), Other |
Other |
1030 |
0.34 |
ADME/T ≤ 10μM |
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 3), Other |
Other |
2700 |
0.31 |
ADME/T ≤ 10μM
|
|
Z81135-2-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #2 Of 6), Other |
Other |
6000 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.49 |
5.45 |
-74.02 |
8 |
6 |
2 |
122 |
342.443 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.68 |
1.99 |
-2.42 |
1 |
1 |
0 |
20 |
114.188 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
4880 |
0.30 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.08 |
9.21 |
-94.07 |
2 |
6 |
0 |
82 |
351.353 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.08 |
7.94 |
-58.12 |
1 |
6 |
-1 |
77 |
350.345 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.66 |
7.04 |
-75.93 |
3 |
6 |
1 |
85 |
352.361 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 32 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
4880 |
0.30 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.08 |
9.21 |
-91.57 |
2 |
6 |
0 |
82 |
351.353 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.08 |
7.97 |
-58.15 |
1 |
6 |
-1 |
77 |
350.345 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.66 |
7.06 |
-75.32 |
3 |
6 |
1 |
85 |
352.361 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 36 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5300 |
0.23 |
Binding ≤ 10μM
|
|
Z100250-1-O |
Calf Thymus DNA (cluster #1 Of 2), Other |
Other |
400 |
0.28 |
Binding ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
9700 |
0.22 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
1259 |
0.26 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
10 |
0.35 |
Functional ≤ 10μM
|
|
Z80018-2-O |
A-375 (Malignant Melanoma Cells) (cluster #2 Of 4), Other |
Other |
26 |
0.33 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
3600 |
0.24 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
6 |
0.36 |
Functional ≤ 10μM
|
|
Z80138-1-O |
G-361 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
1 |
0.39 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
8200 |
0.22 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
66 |
0.31 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
8700 |
0.22 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
7 |
0.36 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
10 |
0.35 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
10 |
0.35 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
9 |
0.35 |
Functional ≤ 10μM
|
|
Z80254-1-O |
MES-SA (Uterine Sarcoma Cells) (cluster #1 Of 2), Other |
Other |
30 |
0.33 |
Functional ≤ 10μM
|
|
Z80255-2-O |
MES-SA/DxS (Uterine Sarcoma Cells) (cluster #2 Of 2), Other |
Other |
28 |
0.33 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
12 |
0.35 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
6 |
0.36 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
4 |
0.37 |
Functional ≤ 10μM
|
|
Z80364-1-O |
P388/ADR (Lymphoma Cells) (cluster #1 Of 1), Other |
Other |
395 |
0.28 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
7 |
0.36 |
Functional ≤ 10μM
|
|
Z80433-1-O |
RPMI-8226 (Multiple Myeloma Cells) (cluster #1 Of 3), Other |
Other |
30 |
0.33 |
Functional ≤ 10μM
|
|
Z80463-1-O |
SCOV-3 (cluster #1 Of 1), Other |
Other |
5 |
0.36 |
Functional ≤ 10μM |
|
Z80475-2-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #2 Of 3), Other |
Other |
16 |
0.34 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
9800 |
0.22 |
Functional ≤ 10μM
|
|
Z80601-1-O |
XRS6 (cluster #1 Of 1), Other |
Other |
10 |
0.35 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
8000 |
0.22 |
Functional ≤ 10μM
|
|
Z80768-1-O |
CH1 (Ovarian Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
3 |
0.37 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
7200 |
0.22 |
Functional ≤ 10μM
|
|
Z81017-2-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
8 |
0.35 |
Functional ≤ 10μM
|
|
Z81034-6-O |
A2780 (Ovarian Carcinoma Cells) (cluster #6 Of 10), Other |
Other |
6 |
0.36 |
Functional ≤ 10μM
|
|
Z81043-1-O |
N592 (cluster #1 Of 2), Other |
Other |
20 |
0.34 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
646 |
0.27 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
100 |
0.31 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
960 |
0.26 |
Functional ≤ 10μM
|
|
Z81261-1-O |
UACC-375 (cluster #1 Of 1), Other |
Other |
48 |
0.32 |
Functional ≤ 10μM
|
|
Z81341-1-O |
Tumor Cell Lines (cluster #1 Of 1), Other |
Other |
27 |
0.33 |
Functional ≤ 10μM
|
|
Q7ZJM1-1-V |
Human Immunodeficiency Virus Type 1 Integrase (cluster #1 Of 6), Viral |
Viruses |
8000 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.36 |
-2.16 |
-97.76 |
10 |
10 |
2 |
172 |
446.504 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.36 |
-3.57 |
-51.37 |
9 |
10 |
1 |
168 |
445.496 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 56 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
-0.4 |
-10.32 |
3 |
4 |
0 |
68 |
137.142 |
1 |
↓
|
|
|
|
Analogs
-
597112
-
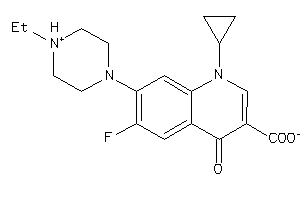
-
6385314
-
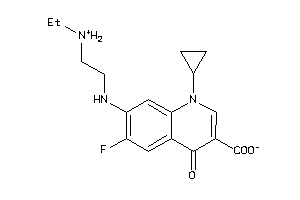
-
39294843
-
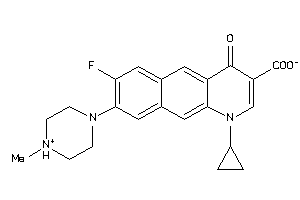
Draw
Identity
99%
90%
80%
70%
Vendors
And 85 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GYRA-1-B |
DNA Gyrase Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
500 |
0.37 |
Binding ≤ 10μM
|
|
GYRB-1-B |
DNA Gyrase Subunit B (cluster #1 Of 1), Bacterial |
Bacteria |
500 |
0.37 |
Binding ≤ 10μM
|
|
PARC-1-B |
Topoisomerase IV Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
2300 |
0.33 |
Binding ≤ 10μM
|
|
GYRA-1-B |
DNA Gyrase Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
200 |
0.39 |
Functional ≤ 10μM
|
|
GYRB-1-B |
DNA Gyrase Subunit B (cluster #1 Of 1), Bacterial |
Bacteria |
200 |
0.39 |
Functional ≤ 10μM
|
|
PARC-1-B |
Topoisomerase IV Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
2500 |
0.33 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3020 |
0.32 |
Binding ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.44 |
Binding ≤ 10μM |
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.44 |
Binding ≤ 10μM |
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
410 |
0.37 |
Binding ≤ 10μM
|
|
Z50117-1-O |
Pseudomonas Aeruginosa (cluster #1 Of 2), Other |
Other |
220 |
0.39 |
Functional ≤ 10μM
|
|
Z50185-3-O |
Staphylococcus Aureus (cluster #3 Of 4), Other |
Other |
400 |
0.37 |
Functional ≤ 10μM
|
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 7), Other |
Other |
10 |
0.47 |
Functional ≤ 10μM
|
|
Z50378-1-O |
Mycobacterium Intracellulare (cluster #1 Of 1), Other |
Other |
910 |
0.35 |
Functional ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
1980 |
0.33 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
9440 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.70 |
9.07 |
-109.14 |
2 |
6 |
0 |
82 |
331.347 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.70 |
7.77 |
-65.4 |
1 |
6 |
-1 |
77 |
330.339 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.45 |
7.06 |
-88.49 |
3 |
6 |
1 |
85 |
332.355 |
2 |
↓
|
|