|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
41 |
0.45 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
80 |
0.43 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM |
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.45 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
11 |
0.48 |
Functional ≤ 10μM
|
|
TRPA1-3-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
230 |
0.40 |
Functional ≤ 10μM |
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.69 |
1.18 |
-5.02 |
1 |
2 |
0 |
29 |
314.469 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.28 |
10.22 |
-11.61 |
2 |
6 |
0 |
80 |
541.866 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 50 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.22 |
12.29 |
-7.95 |
1 |
5 |
0 |
50 |
463.796 |
4 |
↓
|
|
|
|
Analogs
-
13742590
-
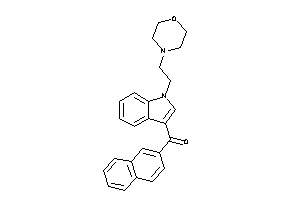
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADCY5-1-E |
Adenylate Cyclase Type V (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
275 |
0.32 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.39 |
Binding ≤ 10μM |
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.39 |
Binding ≤ 10μM |
|
PTGDS-1-E |
Prostaglandin-H2 D-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.40 |
Binding ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
6 |
0.40 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.65 |
11.33 |
-13.19 |
0 |
4 |
0 |
34 |
384.479 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3155 |
0.28 |
Binding ≤ 10μM |
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3155 |
0.28 |
Binding ≤ 10μM |
|
PTGDS-1-E |
Prostaglandin-H2 D-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.27 |
Binding ≤ 10μM |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
453 |
0.32 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
530 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
8.87 |
-10.91 |
0 |
5 |
0 |
44 |
378.472 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
48 |
0.34 |
Binding ≤ 10μM |
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.34 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.12 |
9.14 |
-26.55 |
0 |
5 |
0 |
76 |
453.36 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
-1.2 |
-9.08 |
1 |
6 |
0 |
59 |
459.377 |
5 |
↓
|
|
|
|
Analogs
-
5820249
-
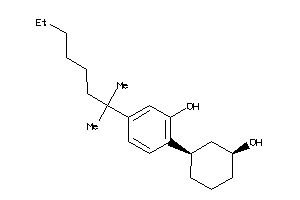
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.46 |
8.08 |
-7.17 |
2 |
2 |
0 |
40 |
318.501 |
7 |
↓
|
|
|
|
Analogs
-
1550857
-
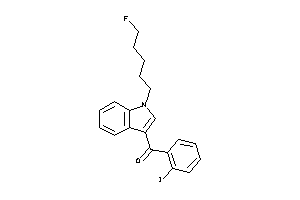
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
560 |
0.34 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
580 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.57 |
14.27 |
-51.78 |
1 |
3 |
1 |
26 |
459.351 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
450 |
0.33 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.15 |
13.84 |
-15.37 |
1 |
3 |
0 |
34 |
356.469 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
17 |
0.44 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
89 |
0.39 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.42 |
Functional ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
24 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.46 |
13.06 |
-16.46 |
0 |
3 |
0 |
31 |
335.447 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
179 |
0.39 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
570 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.87 |
14.41 |
-14.59 |
0 |
2 |
0 |
22 |
319.448 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.72 |
11.14 |
-12.66 |
0 |
5 |
0 |
44 |
426.516 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
11 |
0.45 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
33 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.43 |
13.13 |
-15.54 |
0 |
3 |
0 |
31 |
335.447 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
389 |
0.37 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
499 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.10 |
14.26 |
-14.7 |
0 |
2 |
0 |
22 |
339.866 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
38 |
0.43 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
106 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.08 |
14.25 |
-14.8 |
0 |
2 |
0 |
22 |
339.866 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1064 |
0.33 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
445 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.48 |
13.06 |
-16.17 |
0 |
3 |
0 |
31 |
335.447 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.06 |
14.17 |
-14.03 |
0 |
2 |
0 |
22 |
339.866 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.39 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.20 |
16.42 |
-12.16 |
0 |
2 |
0 |
22 |
385.482 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
16 |
0.38 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.22 |
16.31 |
-10.67 |
0 |
2 |
0 |
22 |
385.482 |
7 |
↓
|
|
|
|
Analogs
-
34326985
-
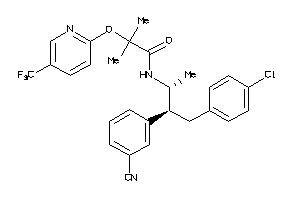
-
34326988
-
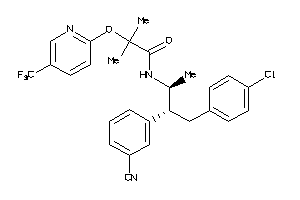
-
34326990
-
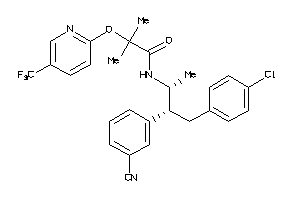
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.52 |
14.96 |
-18.07 |
1 |
5 |
0 |
75 |
515.963 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
15 |
0.39 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
98 |
0.35 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Functional ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
28 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.66 |
15.02 |
-9.44 |
0 |
2 |
0 |
26 |
368.476 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
48 |
0.45 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
179 |
0.41 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
39 |
0.45 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
179 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.65 |
10.07 |
-4.68 |
1 |
2 |
0 |
29 |
314.469 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
390 |
0.33 |
Binding ≤ 10μM
|
|
PTGDS-1-E |
Prostaglandin-H2 D-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
101 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
8.18 |
-14.5 |
0 |
5 |
0 |
44 |
364.445 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
-2.04 |
-7.69 |
1 |
5 |
0 |
50 |
555.247 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.41 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.41 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.94 |
15.64 |
-11.47 |
0 |
2 |
0 |
22 |
355.481 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.87 |
14.08 |
-11.57 |
0 |
2 |
0 |
22 |
327.427 |
4 |
↓
|
|
|
|
Analogs
-
33848576
-
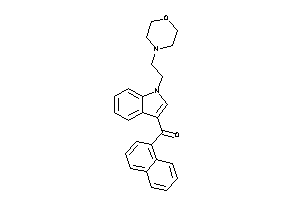
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
59 |
0.35 |
Binding ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
31 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
11.21 |
-10.62 |
0 |
4 |
0 |
34 |
384.479 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
19 |
0.39 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
466 |
0.32 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
466 |
0.32 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.37 |
Functional ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.14 |
-0.41 |
-6.19 |
2 |
3 |
0 |
49 |
386.576 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
856 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.75 |
1.67 |
-6.25 |
0 |
3 |
0 |
30 |
408.76 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
19 |
0.39 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
466 |
0.32 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
150 |
0.34 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
466 |
0.32 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Functional ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
10000 |
0.25 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.14 |
-0.42 |
-6.26 |
2 |
3 |
0 |
49 |
386.576 |
7 |
↓
|
|
|
|
Analogs
-
1550857
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.57 |
14.63 |
-48.58 |
1 |
3 |
1 |
26 |
459.351 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
29 |
0.44 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
146 |
0.40 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.44 |
Functional ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.83 |
14.31 |
-14.49 |
0 |
2 |
0 |
22 |
319.448 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
90 |
0.43 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
159 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.43 |
13.74 |
-14.49 |
0 |
2 |
0 |
22 |
305.421 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
458 |
0.39 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
65 |
0.44 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
146 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.95 |
3.43 |
-3.3 |
0 |
1 |
0 |
9 |
312.497 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
41 |
0.36 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
33 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.24 |
16.31 |
-9.55 |
0 |
2 |
0 |
22 |
385.482 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
11 |
0.38 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.58 |
17.02 |
-9.71 |
0 |
2 |
0 |
22 |
381.519 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
243 |
0.34 |
Binding ≤ 10μM
|
|
CNR1-2-E |
Cannabinoid CB1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
195 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.02 |
13.43 |
-6.53 |
0 |
4 |
0 |
41 |
486.163 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
243 |
0.34 |
Binding ≤ 10μM
|
|
CNR1-2-E |
Cannabinoid CB1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
195 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.02 |
13.42 |
-6.54 |
0 |
4 |
0 |
41 |
486.163 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
466 |
0.32 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
466 |
0.32 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
10000 |
0.25 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.14 |
0.77 |
-7.32 |
2 |
3 |
0 |
49 |
386.576 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
422 |
0.30 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4200 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.56 |
-3.16 |
-8.05 |
1 |
6 |
0 |
59 |
557.219 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
|
KCNH2-3-E |
HERG (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
2700 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.94 |
14.89 |
-7.16 |
1 |
6 |
0 |
59 |
473.795 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4900 |
0.26 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
41 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.88 |
13.53 |
-10.32 |
2 |
3 |
0 |
48 |
400.909 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1070 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.13 |
11.97 |
-9.01 |
0 |
3 |
0 |
24 |
403.353 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.13 |
13.71 |
-52.82 |
1 |
3 |
1 |
25 |
404.361 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
26 |
0.39 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.91 |
15.64 |
-11.58 |
0 |
2 |
0 |
22 |
355.481 |
6 |
↓
|
|
|
|
|
|
|
Analogs
-
1550857
-

-
42966603
-
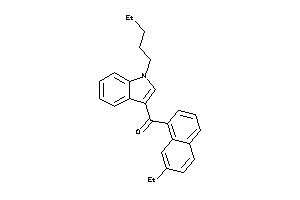
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.58 |
16.42 |
-11.69 |
0 |
2 |
0 |
22 |
369.508 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.75 |
14.46 |
-15.44 |
0 |
3 |
0 |
31 |
371.48 |
7 |
↓
|
|
|
|
Analogs
-
42966603
-

-
1550857
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.11 |
15.69 |
-12.25 |
0 |
2 |
0 |
22 |
355.481 |
6 |
↓
|
|
|
|
|