|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-3-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
|
NMDE3-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.94 |
2.96 |
-76.97 |
10 |
6 |
2 |
130 |
222.296 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
72 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.91 |
2.98 |
-35.87 |
3 |
3 |
1 |
46 |
363.281 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
4200 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
7.78 |
-33.64 |
3 |
2 |
1 |
38 |
245.733 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
970 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
-1.16 |
-35.39 |
3 |
2 |
1 |
37 |
245.733 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AIR5-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.58 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
9100 |
0.39 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
9100 |
0.39 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
9100 |
0.39 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9100 |
0.39 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9100 |
0.39 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
525 |
0.49 |
Binding ≤ 10μM
|
|
NMD3A-3-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
23 |
0.59 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
35 |
0.58 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
23 |
0.59 |
Binding ≤ 10μM
|
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
23 |
0.59 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
23 |
0.59 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
35 |
0.58 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
23 |
0.59 |
Binding ≤ 10μM
|
|
PCP-1-E |
Lysosomal Pro-X Carboxypeptidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
91 |
0.55 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
530 |
0.49 |
Binding ≤ 10μM
|
|
CP2B6-2-E |
Cytochrome P450 2B6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.39 |
ADME/T ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
10 |
0.62 |
Binding ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 1), Other |
Other |
270 |
0.51 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
270 |
0.51 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 3), Other |
Other |
200 |
0.52 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
200 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.82 |
10.57 |
-26.82 |
1 |
1 |
1 |
4 |
244.402 |
2 |
↓
|
|
|
|
Analogs
-
40164019
-
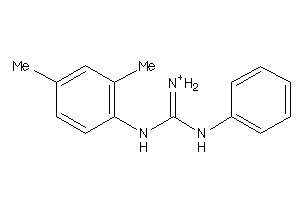
-
40164021
-
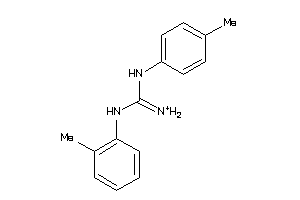
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
744 |
0.48 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2960 |
0.43 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7660 |
0.40 |
Binding ≤ 10μM
|
|
NMD3A-6-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMD3B-6-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE1-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4-6-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
NMDZ1-6-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7800 |
0.40 |
Binding ≤ 10μM
|
|
OPRD-3-E |
Delta Opioid Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3950 |
0.42 |
Binding ≤ 10μM
|
|
OPRK-2-E |
Kappa Opioid Receptor (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3950 |
0.42 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3950 |
0.42 |
Binding ≤ 10μM
|
|
SGMR1-4-E |
Sigma Opioid Receptor (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
89 |
0.55 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9210 |
0.39 |
Functional ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
2000 |
0.44 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
70 |
0.56 |
Binding ≤ 10μM
|
|
Z104302-7-O |
Glutamate NMDA Receptor (cluster #7 Of 7), Other |
Other |
9120 |
0.39 |
Binding ≤ 10μM
|
|
Z50512-3-O |
Cavia Porcellus (cluster #3 Of 7), Other |
Other |
6900 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
8.4 |
-25.53 |
4 |
3 |
1 |
50 |
240.33 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 44 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-6-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
3450 |
0.48 |
Binding ≤ 10μM
|
|
NMD3B-6-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
3450 |
0.48 |
Binding ≤ 10μM
|
|
NMDE1-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3450 |
0.48 |
Binding ≤ 10μM
|
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3450 |
0.48 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3450 |
0.48 |
Binding ≤ 10μM
|
|
NMDE4-6-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
3450 |
0.48 |
Binding ≤ 10μM
|
|
NMDZ1-6-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
3450 |
0.48 |
Binding ≤ 10μM
|
|
SGMR1-4-E |
Sigma Opioid Receptor (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
400 |
0.56 |
Binding ≤ 10μM
|
|
Z104302-7-O |
Glutamate NMDA Receptor (cluster #7 Of 7), Other |
Other |
3450 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
8.37 |
-26.86 |
4 |
3 |
1 |
50 |
212.276 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-6-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
134 |
0.40 |
Binding ≤ 10μM
|
|
NMD3B-6-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
134 |
0.40 |
Binding ≤ 10μM
|
|
NMDE1-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
134 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
134 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
134 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4-6-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
134 |
0.40 |
Binding ≤ 10μM
|
|
NMDZ1-6-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
134 |
0.40 |
Binding ≤ 10μM
|
|
SGMR1-4-E |
Sigma Opioid Receptor (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
165 |
0.40 |
Binding ≤ 10μM
|
|
Z104302-7-O |
Glutamate NMDA Receptor (cluster #7 Of 7), Other |
Other |
330 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.26 |
-0.3 |
-30.1 |
4 |
3 |
1 |
49 |
312.396 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2200 |
0.36 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
2200 |
0.36 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
165 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.41 |
14.12 |
-39.75 |
1 |
1 |
1 |
4 |
294.462 |
6 |
↓
|
|
|
|
Analogs
-
1532560
-
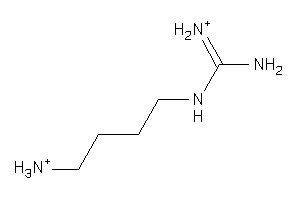
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-1-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
|
NMD3B-1-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
|
NMDE3-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
|
NMDE4-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
|
NMDZ1-5-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.42 |
0.75 |
-75.07 |
10 |
6 |
2 |
127 |
174.252 |
7 |
↓
|
|