|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.04 |
2.11 |
-51.46 |
8 |
13 |
1 |
183 |
482.525 |
9 |
↓
|
|
|
|
Analogs
-
35572125
-
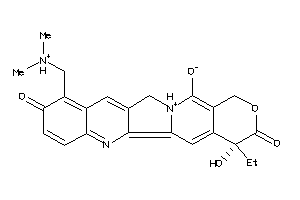
-
35643838
-
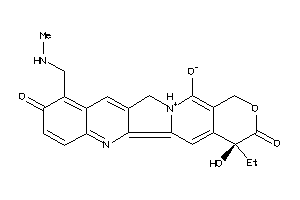
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HIF1A-1-E |
Hypoxia-inducible Factor 1-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.33 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.33 |
Binding ≤ 10μM
|
|
Z100519-1-O |
KETR3 (Renal Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
5 |
0.37 |
Functional ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
13 |
0.36 |
Functional ≤ 10μM
|
|
Z80019-1-O |
A-427 (Lung Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
49 |
0.33 |
Functional ≤ 10μM
|
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
300 |
0.29 |
Functional ≤ 10μM
|
|
Z80039-1-O |
BGC-823 (Stomach Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
3800 |
0.24 |
Functional ≤ 10μM
|
|
Z80105-1-O |
COR-L23 (Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
20 |
0.35 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
1200 |
0.27 |
Functional ≤ 10μM
|
|
Z80145-3-O |
H69 (cluster #3 Of 4), Other |
Other |
63 |
0.33 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
74 |
0.32 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
52 |
0.33 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
880 |
0.27 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
1700 |
0.26 |
Functional ≤ 10μM
|
|
Z80188-1-O |
KB 3-1 (Cervical Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
440 |
0.29 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
601 |
0.28 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
36 |
0.34 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
5550 |
0.24 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
38 |
0.34 |
Functional ≤ 10μM
|
|
Z80316-1-O |
NCI-H128 (cluster #1 Of 1), Other |
Other |
31 |
0.34 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
45 |
0.33 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
45 |
0.33 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
26 |
0.34 |
Functional ≤ 10μM
|
|
Z80482-2-O |
SK-MEL-2 (Melanoma Cells) (cluster #2 Of 4), Other |
Other |
140 |
0.31 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
48 |
0.33 |
Functional ≤ 10μM
|
|
Z80495-1-O |
SK-VLB (cluster #1 Of 1), Other |
Other |
150 |
0.31 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
1500 |
0.26 |
Functional ≤ 10μM
|
|
Z80559-1-O |
U251 (cluster #1 Of 3), Other |
Other |
500 |
0.28 |
Functional ≤ 10μM
|
|
Z80596-1-O |
WISH (Amniotic Epithelial Cells) (cluster #1 Of 1), Other |
Other |
1300 |
0.27 |
Functional ≤ 10μM
|
|
Z80623-1-O |
Panel (56 Tumour Cell Lines) (cluster #1 Of 1), Other |
Other |
160 |
0.31 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
87 |
0.32 |
Functional ≤ 10μM
|
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
78 |
0.32 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
104 |
0.32 |
Functional ≤ 10μM
|
|
Z80799-1-O |
RPMI 8402 (Pre-T-lymphoblastoid Cells) (cluster #1 Of 1), Other |
Other |
21 |
0.35 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
1100 |
0.27 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
56 |
0.33 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
60 |
0.33 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
7100 |
0.23 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
8700 |
0.23 |
Functional ≤ 10μM
|
|
Z81084-1-O |
KB VCR R (cluster #1 Of 1), Other |
Other |
339 |
0.29 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
70 |
0.32 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
9 |
0.36 |
Functional ≤ 10μM
|
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
5 |
0.37 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
79 |
0.32 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
1600 |
0.26 |
Functional ≤ 10μM
|
|
Z81248-1-O |
Malme-3M (Melanoma Cells) (cluster #1 Of 3), Other |
Other |
1000 |
0.27 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
3 |
0.38 |
Functional ≤ 10μM
|
|
Z81342-1-O |
Tumour Cell Lines (cluster #1 Of 1), Other |
Other |
40 |
0.33 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
3100 |
0.25 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
6.39 |
-40.99 |
3 |
8 |
0 |
108 |
422.461 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.31 |
4.23 |
-51.04 |
2 |
8 |
0 |
107 |
421.453 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.31 |
2.6 |
-55.53 |
2 |
8 |
0 |
105 |
421.453 |
3 |
↓
|
|
|
|
Analogs
-
4517652
-
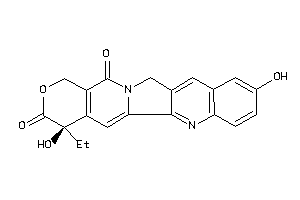
-
4823972
-
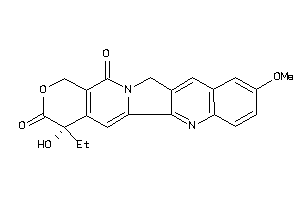
Draw
Identity
99%
90%
80%
70%
Vendors
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.36 |
Binding ≤ 10μM
|
|
Z80099-1-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
9 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
410 |
0.33 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
9 |
0.42 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
4100 |
0.28 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
18 |
0.40 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
12 |
0.41 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
110 |
0.36 |
Functional ≤ 10μM
|
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
280 |
0.34 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
148 |
0.35 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
981 |
0.31 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
700 |
0.32 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
30 |
0.39 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
829 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.53 |
3.91 |
-13.24 |
2 |
7 |
0 |
102 |
364.357 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.28 |
3.5 |
-56.32 |
2 |
7 |
0 |
102 |
364.357 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.28 |
4.82 |
-55.03 |
2 |
7 |
0 |
104 |
364.357 |
1 |
↓
|
|
|
|
Analogs
-
598071
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
840 |
0.32 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.28 |
Binding ≤ 10μM |
|
Z80099-1-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
7 |
0.42 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
7 |
0.42 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
13 |
0.41 |
Functional ≤ 10μM |
|
Z80898-1-O |
NCI-H928 (cluster #1 Of 1), Other |
Other |
8 |
0.42 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
4 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.77 |
4.71 |
-13.32 |
3 |
7 |
0 |
107 |
363.373 |
1 |
↓
|
|
|
|
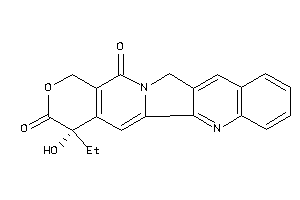
Analogs
-
105309
-
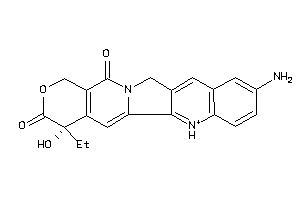
-
1587127
-
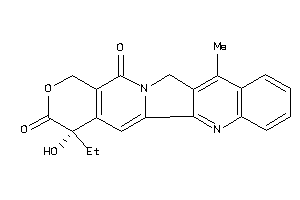
-
22010630
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
6.74 |
-12.85 |
1 |
6 |
0 |
81 |
348.358 |
1 |
↓
|
|
|
|
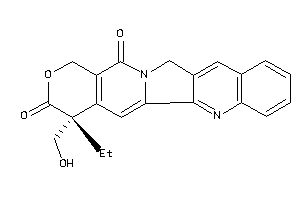
Analogs
-
33822027
-
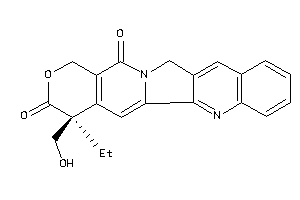
-
33822028
-
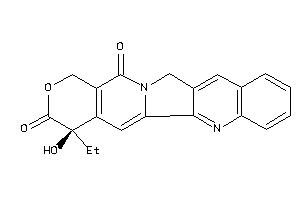
-
1087
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 55 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.31 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.27 |
Binding ≤ 10μM
|
|
CHK1-1-E |
Serine/threonine-protein Kinase Chk1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR1-1-E |
Somatostatin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR2-1-E |
Somatostatin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR3-1-E |
Somatostatin Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR4-1-E |
Somatostatin Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR5-1-E |
Somatostatin Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.40 |
Functional ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.40 |
Functional ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.40 |
Functional ≤ 10μM
|
|
Z100501-1-O |
Lu1 (Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
29 |
0.41 |
Functional ≤ 10μM
|
|
Z100733-1-O |
CT26 (Colon Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
34 |
0.40 |
Functional ≤ 10μM
|
|
Z100752-1-O |
Renca (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
304 |
0.35 |
Functional ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
11 |
0.43 |
Functional ≤ 10μM
|
|
Z103408-1-O |
SW626 (cluster #1 Of 2), Other |
Other |
29 |
0.41 |
Functional ≤ 10μM
|
|
Z103413-1-O |
SW948 (cluster #1 Of 1), Other |
Other |
160 |
0.37 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
794 |
0.33 |
Functional ≤ 10μM
|
|
Z80012-1-O |
A 172 (Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
29 |
0.41 |
Functional ≤ 10μM
|
|
Z80012-1-O |
A 172 (Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
140 |
0.37 |
Functional ≤ 10μM |
|
Z80019-1-O |
A-427 (Lung Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
24 |
0.41 |
Functional ≤ 10μM
|
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
20 |
0.41 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
9550 |
0.27 |
Functional ≤ 10μM |
|
Z80056-1-O |
CAKI-2 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
3960 |
0.29 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
5300 |
0.28 |
Functional ≤ 10μM
|
|
Z80068-3-O |
CCRF-SB (Lymphoblastic Leukemia Cells) (cluster #3 Of 5), Other |
Other |
3 |
0.46 |
Functional ≤ 10μM
|
|
Z80099-1-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
5 |
0.45 |
Functional ≤ 10μM
|
|
Z80115-1-O |
DC3F (cluster #1 Of 1), Other |
Other |
6 |
0.44 |
Functional ≤ 10μM |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
102 |
0.38 |
Functional ≤ 10μM
|
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
210 |
0.36 |
Functional ≤ 10μM |
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
940 |
0.32 |
Functional ≤ 10μM
|
|
Z80145-3-O |
H69 (cluster #3 Of 4), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
7 |
0.44 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
89 |
0.38 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
80 |
0.38 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
90 |
0.38 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
80 |
0.38 |
Functional ≤ 10μM
|
|
Z80188-1-O |
KB 3-1 (Cervical Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
20 |
0.41 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
35 |
0.40 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
60 |
0.39 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
3100 |
0.30 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
7420 |
0.28 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
17 |
0.42 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
4 |
0.45 |
Functional ≤ 10μM
|
|
Z80316-1-O |
NCI-H128 (cluster #1 Of 1), Other |
Other |
18 |
0.42 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z80372-1-O |
P388CPT5 (cluster #1 Of 1), Other |
Other |
2800 |
0.30 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
57 |
0.39 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
1000 |
0.32 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
40 |
0.40 |
Functional ≤ 10μM |
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
20 |
0.41 |
Functional ≤ 10μM
|
|
Z80482-2-O |
SK-MEL-2 (Melanoma Cells) (cluster #2 Of 4), Other |
Other |
7860 |
0.27 |
Functional ≤ 10μM |
|
Z80485-1-O |
SK-MEL-28 (Melanoma Cells) (cluster #1 Of 6), Other |
Other |
40 |
0.40 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
51 |
0.39 |
Functional ≤ 10μM
|
|
Z80495-1-O |
SK-VLB (cluster #1 Of 1), Other |
Other |
53 |
0.39 |
Functional ≤ 10μM
|
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
88 |
0.38 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
600 |
0.34 |
Functional ≤ 10μM
|
|
Z80623-1-O |
Panel (56 Tumour Cell Lines) (cluster #1 Of 1), Other |
Other |
40 |
0.40 |
Functional ≤ 10μM
|
|
Z80647-1-O |
833K Cell Line (cluster #1 Of 1), Other |
Other |
10 |
0.43 |
Functional ≤ 10μM |
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
91 |
0.38 |
Functional ≤ 10μM |
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
7 |
0.44 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
207 |
0.36 |
Functional ≤ 10μM
|
|
Z80751-1-O |
Caov-3 Cell Line (cluster #1 Of 1), Other |
Other |
30 |
0.41 |
Functional ≤ 10μM |
|
Z80751-1-O |
Caov-3 Cell Line (cluster #1 Of 1), Other |
Other |
32 |
0.40 |
Functional ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
45 |
0.40 |
Functional ≤ 10μM
|
|
Z80799-1-O |
RPMI 8402 (Pre-T-lymphoblastoid Cells) (cluster #1 Of 1), Other |
Other |
6 |
0.44 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80898-1-O |
NCI-H928 (cluster #1 Of 1), Other |
Other |
8 |
0.44 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z80930-1-O |
HEC-1B Cell Line (cluster #1 Of 1), Other |
Other |
60 |
0.39 |
Functional ≤ 10μM
|
|
Z80930-1-O |
HEC-1B Cell Line (cluster #1 Of 1), Other |
Other |
8640 |
0.27 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
17 |
0.42 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
60 |
0.39 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
330 |
0.35 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
7 |
0.44 |
Functional ≤ 10μM
|
|
Z81054-1-O |
SF-268 (Glioblastoma Cells) (cluster #1 Of 3), Other |
Other |
59 |
0.39 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
6 |
0.44 |
Functional ≤ 10μM
|
|
Z81080-1-O |
KATO-III Cell Line (cluster #1 Of 1), Other |
Other |
448 |
0.34 |
Functional ≤ 10μM
|
|
Z81080-1-O |
KATO-III Cell Line (cluster #1 Of 1), Other |
Other |
1170 |
0.32 |
Functional ≤ 10μM |
|
Z81084-1-O |
KB VCR R (cluster #1 Of 1), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z81134-1-O |
L2987 (Lung Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
400 |
0.34 |
Functional ≤ 10μM
|
|
Z81160-1-O |
Lewis Lung Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
33 |
0.40 |
Functional ≤ 10μM
|
|
Z81164-1-O |
LL Cell Line (cluster #1 Of 2), Other |
Other |
33 |
0.40 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
86 |
0.38 |
Functional ≤ 10μM
|
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
48 |
0.39 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
710 |
0.33 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
940 |
0.32 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
970 |
0.32 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
2 |
0.47 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
83 |
0.38 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
890 |
0.33 |
ADME/T ≤ 10μM |
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
500 |
0.34 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
6.76 |
-12.81 |
1 |
6 |
0 |
81 |
348.358 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
5.41 |
-13.93 |
1 |
8 |
0 |
100 |
392.367 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.89 |
5.66 |
-45.69 |
2 |
8 |
1 |
101 |
393.375 |
1 |
↓
|
|
|
|
Analogs
-
37866089
-
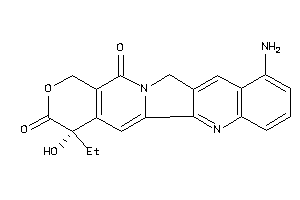
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
840 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.77 |
4.68 |
-13.45 |
3 |
7 |
0 |
107 |
363.373 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.44 |
6.68 |
-52.29 |
2 |
10 |
1 |
108 |
519.578 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.44 |
6.56 |
-57.91 |
2 |
10 |
1 |
108 |
519.578 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.44 |
4.21 |
-14.26 |
1 |
10 |
0 |
106 |
518.57 |
3 |
↓
|
|
|
|
Analogs
-
1587126
-
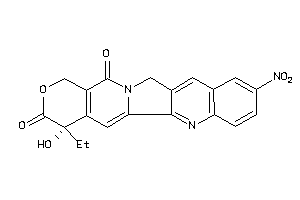
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
640 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.96 |
7.4 |
-14.46 |
1 |
9 |
0 |
127 |
393.355 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.96 |
7.65 |
-54.81 |
2 |
9 |
1 |
128 |
394.363 |
2 |
↓
|
|
|
|
Analogs
-
538323
-
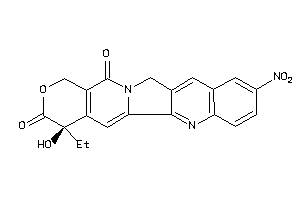
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
640 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.96 |
-1.4 |
-14.46 |
1 |
9 |
0 |
127 |
393.355 |
2 |
↓
|
|
|
|
Analogs
-
33822027
-

-
33822028
-

-
1087
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
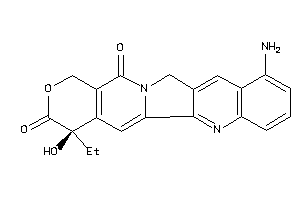
(4S)-9-Amino-4-ethyl-4-hydroxy-1H-pyrano(3',4':6,7)indolizino(1,2-b)quinoline-3,14(4H,12H)-dione; 10-Aminocamptothecin; 1H-Pyrano(3',4':6,7)indolizino(1,2-b)quinoline-3,14(4H,12H)-dione, 9-amino-4-ethyl-4-hydroxy-, (S)-; 9-amino-20(S)-camptothecin; 9-amin
(4S)-9-Amino-4-ethyl-4-hydroxy-1…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
140 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.08 |
-3.96 |
-13.12 |
3 |
7 |
0 |
107 |
363.373 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.08 |
-3.91 |
-41.37 |
4 |
7 |
1 |
108 |
364.381 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
140 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
7.24 |
-11.55 |
1 |
6 |
0 |
81 |
382.803 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.68 |
7.49 |
-44.92 |
2 |
6 |
1 |
83 |
383.811 |
1 |
↓
|
|
|
|
Analogs
-
13000556
-
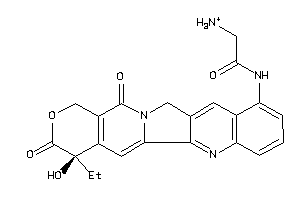
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.63 |
3.3 |
-65.97 |
5 |
9 |
1 |
138 |
421.433 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.63 |
2.9 |
-17.51 |
4 |
9 |
0 |
137 |
420.425 |
3 |
↓
|
|
|
|
Analogs
-
1625746
-
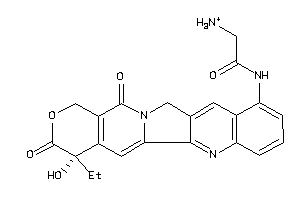
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.63 |
3.27 |
-65.95 |
5 |
9 |
1 |
138 |
421.433 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.63 |
3.52 |
-113.19 |
6 |
9 |
2 |
139 |
422.441 |
3 |
↓
|
|
|
|
Analogs
-
967733
-
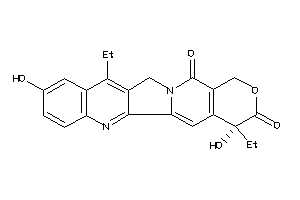
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.26 |
Binding ≤ 10μM
|
|
Z80014-1-O |
A204 (Rhabdomyosarcoma Cells) (cluster #1 Of 1), Other |
Other |
20 |
0.37 |
Functional ≤ 10μM
|
|
Z80019-1-O |
A-427 (Lung Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
1 |
0.43 |
Functional ≤ 10μM
|
|
Z80021-2-O |
A498 (Renal Carcinoma Cells) (cluster #2 Of 5), Other |
Other |
126 |
0.33 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
5 |
0.40 |
Functional ≤ 10μM
|
|
Z80045-1-O |
BT-20 (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
316 |
0.31 |
Functional ≤ 10μM
|
|
Z80048-1-O |
BXPC-3 (Pancreatic Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
79 |
0.34 |
Functional ≤ 10μM
|
|
Z80056-1-O |
CAKI-2 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
5012 |
0.26 |
Functional ≤ 10μM
|
|
Z80086-1-O |
CFPAC-1 (Pancreatic Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
5 |
0.40 |
Functional ≤ 10μM
|
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
158 |
0.33 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
4 |
0.41 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
19 |
0.37 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
670 |
0.30 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
9 |
0.39 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
398 |
0.31 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
370 |
0.31 |
Functional ≤ 10μM
|
|
Z80254-1-O |
MES-SA (Uterine Sarcoma Cells) (cluster #1 Of 2), Other |
Other |
79 |
0.34 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
8 |
0.39 |
Functional ≤ 10μM
|
|
Z80316-1-O |
NCI-H128 (cluster #1 Of 1), Other |
Other |
6 |
0.40 |
Functional ≤ 10μM
|
|
Z80381-1-O |
PANC-1 (Pancreatic Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
7943 |
0.25 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
36 |
0.36 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
4 |
0.41 |
Functional ≤ 10μM
|
|
Z80485-1-O |
SK-MEL-28 (Melanoma Cells) (cluster #1 Of 6), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
720 |
0.30 |
Functional ≤ 10μM
|
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
6 |
0.40 |
Functional ≤ 10μM
|
|
Z80539-1-O |
T98G (Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
1 |
0.43 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
8 |
0.39 |
Functional ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
40 |
0.36 |
Functional ≤ 10μM
|
|
Z80640-1-O |
786-0 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
5 |
0.40 |
Functional ≤ 10μM
|
|
Z80661-1-O |
A704 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
10000 |
0.24 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
88 |
0.34 |
Functional ≤ 10μM
|
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
3190 |
0.27 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
50 |
0.35 |
Functional ≤ 10μM
|
|
Z80800-1-O |
MIA PaCa-2 (Pancreatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
251 |
0.32 |
Functional ≤ 10μM
|
|
Z80847-1-O |
FaDu (Pharyngeal Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
1 |
0.43 |
Functional ≤ 10μM
|
|
Z80891-1-O |
H4 (Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
316 |
0.31 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
7 |
0.39 |
Functional ≤ 10μM
|
|
Z80991-1-O |
Hs-578T (Breast Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
501 |
0.30 |
Functional ≤ 10μM
|
|
Z80996-1-O |
HT1197 (Bladder Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
10000 |
0.24 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
251 |
0.32 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
80 |
0.34 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
3162 |
0.27 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
39 |
0.36 |
Functional ≤ 10μM
|
|
Z81248-1-O |
Malme-3M (Melanoma Cells) (cluster #1 Of 3), Other |
Other |
6310 |
0.25 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
5012 |
0.26 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
79 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
5.17 |
-12.57 |
2 |
7 |
0 |
102 |
392.411 |
2 |
↓
|
|
|
|
Analogs
-
13284239
-
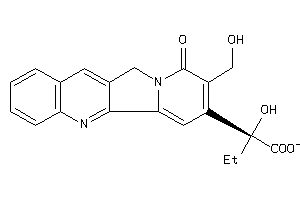
-
16250
-
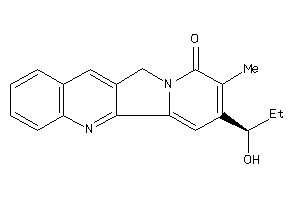
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.30 |
Binding ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
14 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.92 |
5.2 |
-56.27 |
2 |
7 |
-1 |
115 |
365.365 |
4 |
↓
|
|
|
|
Analogs
-
16250
-

-
3954245
-
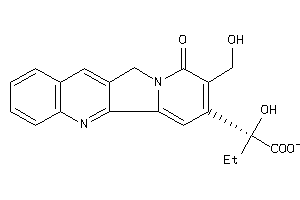
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.92 |
3.01 |
-65.36 |
2 |
7 |
-1 |
115 |
365.365 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.92 |
3.26 |
-65.84 |
3 |
7 |
0 |
117 |
366.373 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.34 |
6.11 |
-12.21 |
1 |
7 |
0 |
91 |
378.384 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.34 |
6.36 |
-40.34 |
2 |
7 |
1 |
92 |
379.392 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.34 |
6.09 |
-12.26 |
1 |
7 |
0 |
91 |
378.384 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.34 |
6.34 |
-40.4 |
2 |
7 |
1 |
92 |
379.392 |
2 |
↓
|
|
|
|
Analogs
-
13832891
-
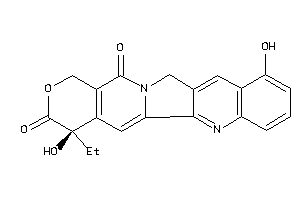
-
26891987
-
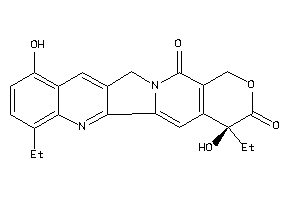
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
3.97 |
-12.98 |
2 |
7 |
0 |
102 |
364.357 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.06 |
4.75 |
-41.09 |
1 |
7 |
-1 |
104 |
363.349 |
1 |
↓
|
|
|
|
Analogs
-
5924106
-
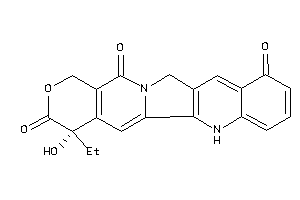
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
3.95 |
-13.18 |
2 |
7 |
0 |
102 |
364.357 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.06 |
4.2 |
-42.97 |
3 |
7 |
1 |
103 |
365.365 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABI-1-B |
Enoyl-[acyl-carrier-protein] Reductase (cluster #1 Of 2), Bacterial |
Bacteria |
7050 |
0.34 |
Binding ≤ 10μM
|
|
NANH-1-B |
Sialidase (cluster #1 Of 1), Bacterial |
Bacteria |
4300 |
0.36 |
Binding ≤ 10μM
|
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1660 |
0.39 |
Binding ≤ 10μM
|
|
ABCG2-1-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8900 |
0.34 |
Binding ≤ 10μM
|
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
9570 |
0.33 |
Binding ≤ 10μM
|
|
AOFA-4-E |
Monoamine Oxidase A (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
4900 |
0.35 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CD38-1-E |
Lymphocyte Differentiation Antigen CD38 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8200 |
0.34 |
Binding ≤ 10μM |
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
79 |
0.47 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.41 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
800 |
0.41 |
Binding ≤ 10μM
|
|
LGUL-2-E |
Glyoxalase I (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7700 |
0.34 |
Binding ≤ 10μM
|
|
LOX1-1-E |
Seed Lipoxygenase-1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.37 |
Binding ≤ 10μM
|
|
NOX4-1-E |
NADPH Oxidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
850 |
0.40 |
Binding ≤ 10μM
|
|
Q965D5-1-E |
Enoyl-acyl-carrier Protein Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.38 |
Binding ≤ 10μM
|
|
Q965D6-1-E |
3-oxoacyl-acyl-carrier Protein Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.41 |
Binding ≤ 10μM
|
|
Q965D7-2-E |
Fatty Acid Synthase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.35 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.41 |
Binding ≤ 10μM
|
|
XDH-2-E |
Xanthine Dehydrogenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1450 |
0.39 |
Functional ≤ 10μM
|
|
CP1A1-1-E |
Cytochrome P450 1A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
890 |
0.40 |
ADME/T ≤ 10μM
|
|
CP1A2-1-E |
Cytochrome P450 1A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3370 |
0.36 |
ADME/T ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
56 |
0.48 |
ADME/T ≤ 10μM
|
|
Z104294-2-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #2 Of 2), Other |
Other |
3800 |
0.36 |
Binding ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
9600 |
0.33 |
Functional ≤ 10μM
|
|
Z50607-4-O |
Human Immunodeficiency Virus 1 (cluster #4 Of 10), Other |
Other |
10000 |
0.33 |
Functional ≤ 10μM |
|
Z50652-3-O |
Influenza A Virus (cluster #3 Of 4), Other |
Other |
6820 |
0.34 |
Functional ≤ 10μM
|
|
Z80150-2-O |
H9c2 (Cardiomyoblast Cells) (cluster #2 Of 2), Other |
Other |
5530 |
0.35 |
Functional ≤ 10μM
|
|
Z80418-2-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #2 Of 9), Other |
Other |
7500 |
0.34 |
Functional ≤ 10μM
|
|
Z80420-1-O |
RBL-2H3 (Basophilic Leukemia Cells) (cluster #1 Of 2), Other |
Other |
5800 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
-0.94 |
-14.61 |
4 |
6 |
0 |
111 |
286.239 |
1 |
↓
|
|