|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
2.17 |
-7.29 |
1 |
3 |
0 |
42 |
181.166 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.65 |
1.84 |
-3.95 |
0 |
2 |
0 |
26 |
166.077 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.21 |
4.71 |
-34.24 |
1 |
2 |
1 |
23 |
164.228 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
3.97 |
-4.87 |
1 |
2 |
0 |
33 |
226.073 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
3.59 |
-6.86 |
1 |
3 |
0 |
42 |
193.246 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
3.84 |
-34.68 |
1 |
2 |
1 |
23 |
150.201 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.29 |
7.07 |
-3.43 |
0 |
0 |
0 |
0 |
286.034 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
8.26 |
-6.11 |
0 |
2 |
0 |
26 |
271.154 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
39564223
-
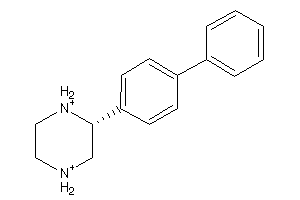
-
39564224
-
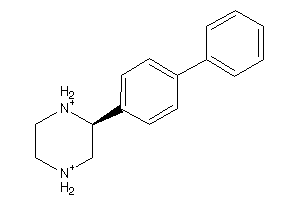
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
4.5 |
-120.6 |
4 |
2 |
2 |
33 |
178.279 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.42 |
3.16 |
-35.02 |
3 |
2 |
1 |
29 |
177.271 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.42 |
2.04 |
-2.21 |
2 |
2 |
0 |
24 |
176.263 |
1 |
↓
|
|
|
|
Analogs
-
39295281
-
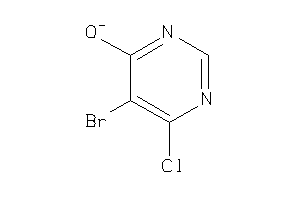
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
-0.49 |
-36.92 |
0 |
3 |
-1 |
49 |
129.526 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.48 |
0.61 |
-15.3 |
1 |
3 |
0 |
46 |
130.534 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.95 |
2.3 |
-30.64 |
0 |
5 |
-1 |
82 |
139.09 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.95 |
1.57 |
-10.71 |
1 |
5 |
0 |
79 |
140.098 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.95 |
1.8 |
-37.83 |
2 |
5 |
1 |
80 |
141.106 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
3.84 |
-34.05 |
0 |
2 |
-1 |
47 |
186.112 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.50 |
3.07 |
-7.11 |
1 |
2 |
0 |
44 |
187.12 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.35 |
1.63 |
-41.26 |
2 |
3 |
0 |
57 |
151.112 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.35 |
0.38 |
-43.82 |
1 |
3 |
-1 |
52 |
150.104 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-5-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #5 Of 12), Other |
Other |
1210 |
0.69 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156 |
Z80156
|
HL-60 (Promyeloblast Leukemia Cells) |
1210 |
0.69 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
6.74 |
-46.91 |
3 |
7 |
1 |
75 |
428.553 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.92 |
4.54 |
-14.78 |
2 |
7 |
0 |
74 |
427.545 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.92 |
6.89 |
-46.27 |
3 |
7 |
1 |
75 |
428.553 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
6.81 |
-48.71 |
3 |
7 |
1 |
75 |
428.553 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.92 |
6.92 |
-48.11 |
3 |
7 |
1 |
75 |
428.553 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.92 |
4.61 |
-16.16 |
2 |
7 |
0 |
74 |
427.545 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
-0.04 |
7.83 |
-63.43 |
1 |
7 |
-1 |
87 |
374.392 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.04 |
7.05 |
-52.32 |
3 |
7 |
1 |
88 |
376.408 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.78 |
7.73 |
-77.5 |
3 |
7 |
1 |
94 |
376.408 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
6.74 |
-29.19 |
3 |
3 |
1 |
45 |
216.308 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.79 |
6.28 |
-8.91 |
2 |
3 |
0 |
44 |
215.3 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
34219725
-
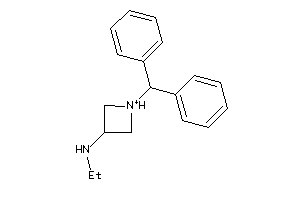
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.24 |
8.11 |
-126.36 |
4 |
2 |
2 |
32 |
240.35 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.24 |
5.14 |
-3.73 |
2 |
2 |
0 |
29 |
238.334 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.24 |
5.59 |
-43.57 |
3 |
2 |
1 |
31 |
239.342 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
1.97 |
-17.33 |
1 |
2 |
0 |
37 |
208.442 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.40 |
0.72 |
-7.53 |
1 |
2 |
0 |
33 |
208.442 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.40 |
1.51 |
-32.02 |
0 |
2 |
-1 |
36 |
207.434 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.31 |
4.18 |
-7.77 |
0 |
4 |
0 |
56 |
265.696 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.31 |
3.54 |
-8.62 |
0 |
4 |
0 |
56 |
265.696 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.90 |
2.13 |
-6.88 |
1 |
3 |
0 |
46 |
178.113 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.36 |
1.16 |
-35.51 |
0 |
3 |
-1 |
49 |
177.105 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.90 |
1.58 |
-16.49 |
1 |
3 |
0 |
46 |
178.113 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.08 |
1.39 |
-104.11 |
4 |
2 |
2 |
33 |
116.208 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.08 |
0.15 |
-35.27 |
3 |
2 |
1 |
29 |
115.2 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.08 |
0.04 |
-35.21 |
3 |
2 |
1 |
29 |
115.2 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.08 |
1.39 |
-104.07 |
4 |
2 |
2 |
33 |
116.208 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.08 |
0.12 |
-36.88 |
3 |
2 |
1 |
29 |
115.2 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.08 |
0.04 |
-34.72 |
3 |
2 |
1 |
29 |
115.2 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
14.52 |
-62.52 |
1 |
5 |
1 |
53 |
424.471 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.10 |
12.26 |
-9.58 |
0 |
5 |
0 |
52 |
423.463 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.10 |
13.87 |
-55.14 |
1 |
5 |
1 |
53 |
424.471 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
10 |
-52.97 |
1 |
5 |
1 |
53 |
312.393 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.99 |
10.06 |
-47.08 |
1 |
5 |
1 |
53 |
312.393 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.99 |
7.68 |
-7.33 |
0 |
5 |
0 |
52 |
311.385 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.13 |
6 |
-129.26 |
4 |
2 |
2 |
33 |
214.312 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.13 |
3.36 |
-3.77 |
2 |
2 |
0 |
24 |
212.296 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.13 |
4.66 |
-40.05 |
3 |
2 |
1 |
29 |
213.304 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.13 |
5.96 |
-129.41 |
4 |
2 |
2 |
33 |
214.312 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.13 |
3.62 |
-3.58 |
2 |
2 |
0 |
24 |
212.296 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.13 |
4.63 |
-38.49 |
3 |
2 |
1 |
29 |
213.304 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
4.83 |
-39.11 |
0 |
4 |
-1 |
69 |
216.998 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.41 |
3.8 |
-10.66 |
1 |
4 |
0 |
66 |
218.006 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.75 |
1.72 |
-29.42 |
0 |
4 |
-1 |
64 |
93.069 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
-0.75 |
1.74 |
-29.33 |
0 |
4 |
-1 |
64 |
93.069 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
-0.75 |
1.83 |
-29 |
0 |
4 |
-1 |
64 |
93.069 |
0 |
↓
|
|
|
|
Analogs
-
39134315
-
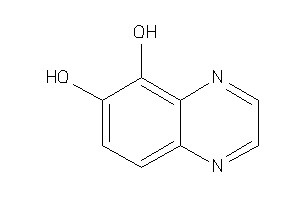
-
39248924
-
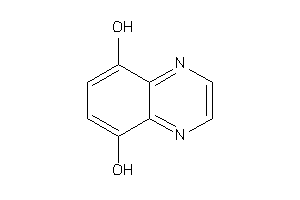
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
2.04 |
-39.94 |
0 |
3 |
-1 |
49 |
145.141 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.47 |
1.19 |
-8.97 |
1 |
3 |
0 |
46 |
146.149 |
0 |
↓
|
|
|
|
Analogs
-
21298676
-
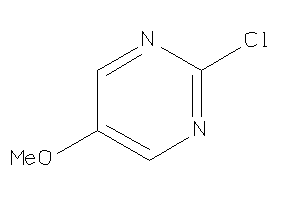
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
0.26 |
-31.84 |
0 |
3 |
-1 |
49 |
129.526 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.94 |
-0.51 |
-6.34 |
1 |
3 |
0 |
46 |
130.534 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
4.86 |
-127.42 |
4 |
2 |
2 |
33 |
251.132 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.35 |
2.21 |
-2.22 |
2 |
2 |
0 |
24 |
249.116 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.35 |
3.56 |
-42.23 |
3 |
2 |
1 |
29 |
250.124 |
1 |
↓
|
|