|
|
Analogs
-
3776592
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102352-3-O |
Plasma (cluster #3 Of 3), Other |
Other |
17 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.17 |
7.38 |
-56.8 |
2 |
5 |
-1 |
90 |
397.491 |
8 |
↓
|
|
|
|
Analogs
-
3776592
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102352-3-O |
Plasma (cluster #3 Of 3), Other |
Other |
17 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.17 |
7.72 |
-56.81 |
2 |
5 |
-1 |
90 |
397.491 |
8 |
↓
|
|
|
|
Analogs
-
39392275
-
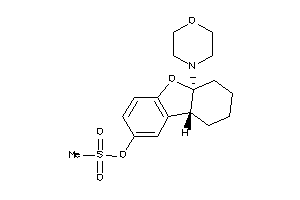
-
39392277
-
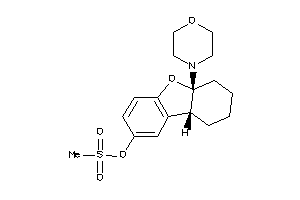
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.10 |
1.83 |
-6.65 |
1 |
4 |
0 |
42 |
261.321 |
1 |
↓
|
|
|
|
Analogs
-
39392275
-

-
39392277
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.10 |
0.89 |
-6.43 |
1 |
4 |
0 |
42 |
261.321 |
1 |
↓
|
|
|
|
Analogs
-
39392275
-

-
39392277
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.10 |
0.87 |
-9.15 |
1 |
4 |
0 |
42 |
261.321 |
1 |
↓
|
|
|
|
Analogs
-
39392275
-

-
39392277
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.10 |
1.82 |
-8.98 |
1 |
4 |
0 |
42 |
261.321 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
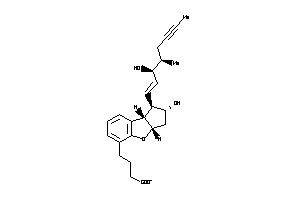
Popular Name:
4-[(1S,2S,3aR,8bR)-2-hydroxy-1-[(E,3R,4R)-3-hydroxy-4-methyl-oct-1-en-6-ynyl]-2,3,3a,8b-tetrahydro-1
4-[(1S,2S,3aR,8bR)-2-hydroxy-1-[…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.17 |
8.61 |
-58.21 |
2 |
5 |
-1 |
90 |
397.491 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.17 |
6.63 |
-15.33 |
3 |
5 |
0 |
87 |
398.499 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
4-[(1S,2S,3aR,8bR)-2-hydroxy-1-[(E,3R,4S)-3-hydroxy-4-methyl-oct-1-en-6-ynyl]-2,3,3a,8b-tetrahydro-1
4-[(1S,2S,3aR,8bR)-2-hydroxy-1-[…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.17 |
8.76 |
-57.94 |
2 |
5 |
-1 |
90 |
397.491 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.17 |
6.78 |
-15.28 |
3 |
5 |
0 |
87 |
398.499 |
8 |
↓
|
|